Int J App Pharm, Vol 17, Issue 2, 2025, 134-141Original Article
IN SILICO STUDIES AND CYTOTOXICITY ASSAY OF BENZYLIDENE BENZO HYDRAZIDE DERIVATIVES ON CANCER STEM CELL
IMANUEL GAURU1,2, YUSUF S. ALAM1, MARDI SANTOSO1, ARIF FADLAN1, NUR R. AFFIFAH1, VINDA A. N. ANDIFA3, PRATIWI PUDJIASTUTI4, FAHIMAH MARTAK1*
1Department of Chemistry, Faculty of Science and Data Analytics, Sepuluh Nopember Institute of Technology, Surabaya-60111, Indonesia. 2Department of Chemistry, Faculty of Science and Engineering, University of Nusa Cendana, Indonesia. 3Department of Biology, Faculty of Science and Data Analytics, Sepuluh Nopember Institute of Technology, Surabaya-60111, Indonesia. 4Department of Chemistry, Faculty of Science and Technology, Airlangga University, Indonesia
*Corresponding author: Fahimah Martak; *Email: fahimahm@chem.its.ac.id
Received: 04 Nov 2024, Revised and Accepted: 14 Jan 2025
ABSTRACT
Objective: This study aimed to evaluate the biological activity of benzylidene benzohydrazide derivatives against Cancer Stem Cells (CSCs) through in vitro cytotoxicity tests and silico analyses using molecular docking.
Methods: Four hydrazone compounds, namely benzylidene benzo hydrazide (L1), 2-methyl benzylidene benzo hydrazide (L2), 2-nitro benzylidene benzo hydrazide (L3), and 2-bromobenzylidene benzo hydrazide (L4) were used for in silico and in vitro studies. The interaction between hydrazone compounds and the EGFR protein receptor (PDB ID: 1m17) was investigated using the AutoDock tools 1.5.7. The PASS server predicted the biological activities of hydrazone substances. ADMET of hydrazone compounds was assessed using the ADMETLab 2.0. Meanwhile, the cytotoxic activity test of hydrazone compounds on CSCs was evaluated using the MTT Assay method.
Results: The results of molecular docking analysis of test compounds L1-L4 provide binding energy values ranging from -6.69 to-7.74 kcal/mol. The binding energy value of L1-L4 is lower than the reference Doxorubicin (-4.30 Kcal/mol). The results of the cytotoxicity test of test compounds with CSCs provide IC50 results for L1 of 0.220±0.360 μg/ml, L2 of 0.034±0.023 μg/ml, L3 of 0.355±0.276 μg/ml, L4 of 1.193±1.122 μg/ml and Doxorubicin of 0.220±0.180 μg/ml. These results indicate that hydrazone derivatives have the potential to be CSCs inhibitor.
Conclusion: 2-methyl benzylidene benzo hydrazide (L2) had the potential as a CSCs inhibitor with vigorous cytotoxic activity in vitro against CSCs cell lines
Keywords: Hydrazone, In silico, Cytotoxicity, Cancer stem cells, Drug-likeness
© 2025 The Authors. Published by Innovare Academic Sciences Pvt Ltd. This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/)
DOI: https://dx.doi.org/10.22159/ijap.2025v17i2.53105 Journal homepage: https://innovareacademics.in/journals/index.php/ijap
INTRODUCTION
Cancer is one of the most devastating illnesses affecting populations globally despite advances in various therapeutic interventions. A significant problem commonly associated with cancer treatment is drug resistance. This phenomenon has resulted in extensive studies on Cancer Stem Cells (CSCs), establishing its close relationship with various aspects of tumorigenesis, including progression, metastasis, recurrence, and therapeutic resistance, potential causes of tumor treatment failure [1]. CSCs arising from normal stem cells with oncogene mutations plays a role in tumor formation, development, metastasis, treatment resistance, recurrence, and unfortunate prognosis [2]. These normal stem cells transform into tumor cells, which have metastatic properties and are invasive in the new environment. Furthermore, their progenitors mature and become tumor cells [3]. In several studies, the level of EGFR (Epidermal Growth Factor Receptor) in CSCs is related to unfavorable prognosis and resistance to cancer treatment [4, 5]. Most of their functions depend on EGFR, including stemness, metabolism, immune modulatory activity, dormancy, and resistance to therapy [6, 7]. Various hydrazone substances have also been made and evaluated as blockers of critical cellular pathways, including EGFR. For example, the 4-amino-6-arylaminopyrimidine-5-carbaldehyde hydrazone compounds have excellent ErbB-2/EGFR tyrosine kinase inhibition, with potential as ErbB-2/EGFR dual kinase inhibitors [8]. Therefore, there is a need to develop new, diverse, and multi-targeted treatment methods due to the distinct characteristics of cancer cells that remain unexplored. The hydrazones used in this study result from synthesizing benzaldehyde and benzohydrazide. Benzaldehyde was chosen because it contains a carbonyl group (C=O), which is very reactive to nucleophilic groups such as amine groups (-NH₂) in benzohydrazide. This allows the condensation reaction to form a hydrazone bond (-C=N-NH-). Hydrazones formed from benzaldehyde derivatives tend to be stable because the aromatic ring system in benzaldehyde provides resonance stabilization. In addition, the hydrazide group also increases the stability of the product through the possibility of intra-or inter-molecular hydrogen interactions. Hydrazones derived from benzaldehyde and benzohydrazide can target specific pathways essential for CSCs survival, such as the Notch, Hedgehog, and Wnt pathways involved in CSCs self-renewal [9]. Gold nanoparticles bound to doxorubicin via poly(ethylene glycol) spacer and hydrazone bond (DOX-Hyd@AuNPs) were able to mediate potent delivery of doxorubicin to breast CSCs and reduce their mammosphere-forming capacity and cancer initiation activity, leading to marked enhancement of tumor growth inhibition in murine models. Nanoparticle (DOX-Hyd@AuNPs)-mediated drug delivery also markedly attenuated tumor growth during the off-therapy phase by reducing breast CSCs in the tumor [10]. Several studies show hydrazone has various pharmacological activities ranging from anticancer, antiviral, antibacterial, anti-inflammatory, antiprotozoal, antioxidant, antihypertensive, anticonvulsant, antimalarial, antifungal, to being a heart medicine [11–14]. Hydrazone compounds can also act as an androgen receptor inhibitor in "LnCaP" prostate cancer cells [15]. Moreover, derivatives from salicylaldehyde and hydrazide provided bioactivity (IC50) in lung "A549" and breast cancer cell "MCF-7" of 47.36±3.81 µM and 18.61±2.18 µM, respectively [16], which is included in the moderate toxicity category. Replacement of the methyl substituent on the hydrazide with a phenyl group (R2 = –CH3 → –Ph) has shown an increasing trend in the cytotoxicity of compounds to 5.27±0.45 µM and 3.66±0.38 µM. This indicates that hydrazone compounds can be an alternative anticancer agent. In silico studies using molecular docking methods are an important initial part of drug discovery, design, and development [17]. This method has several advantages, including reducing excessive equipment and materials and saving experimental costs [18]. In silico analyses can also be employed to forecast the performances of hydrazone sub-stances by estimating the bond-free energy generated during the interaction with the active site of the protein involved [19]. In silico combined with in vitro methods has exhibited numerous benefits in studying the structure, properties, and biological activity of compounds with the potential as drugs [20]. Multiple investigations have been carried out to identify new agents targeting CSCs. Still, there are no prior studies on in silico and in vitro analyses of benzylidene benzohydrazide derivatives substances as CSCs inhibitors. Consequently, this aimed to evaluate the potential of four hydrazone compounds in CSCs inhibition by testing their in vitro toxicity properties and in silico interactions.
MATERIALS AND METHODS
Materials
Here, all four hydrazone compounds were synthesized, purified, and characterized following the reported procedure by Yusuf et al. (2023) [21], and their structures are shown in table 2. The software employed was Chemdraw Ultra 12.0 (cambridgesoft), Discovery Studio Visualizer (BIOVIA), MGL Tools 1.5.7, and AutoDock Tools 1.5.7. The proteins analyzed were Epidermal Growth Factor Receptor (EGFR) tyrosine kinase, obtained from the RSCB database, with a PDB ID 1m17 [22]. The materials needed for the cytotoxicity assay included a conical tube, 96-well plate, micropipette, tip, microtube, Biological Safety Cabinet (BSC), centrifuge, inverted microscope, hemocytometer, ELISA reader, cryotube, liquid nitrogen tank, incubator, Cancer Stem Cells (CSCs) from the SCCR Laboratory (Stem Cell and Cancer Research Center) Sultan Agung University (Unissula), Medium Dulbecco's Modified Eeagle's Medium (DMEM) F12, Penicillin-streptomycin 1.5%, Fungizone (Amphotericin B) 0.25%, Fetal Bovine Serum (FBS) 10%, Phosphate Buffered Saline (PBS); Trypsin-EDTA 0.25%, Dimethyl Sulfoxide (DMSO) solvent, Sodium Dodecyl Sulfate reagent (SDS-stopper), MTT solution, doxorubicnin, and 70% alcohol.
Anticancer profile prediction
The prediction of the biological activities of hydrazone substances was performed by the Prediction of Activity Spectra for Substances (PASS) server, retrieved via the website http://www.way2drug.com/ PASSOnline/predict.php. PASS prediction showed the predicted activity spectrum based on probable activity and inactivity, represented by Pa and Pi, respectively. In this study, the biological activity analyzed was the potential of hydrazone compounds as an anticancer. The results showed various probabilities of Pa and Pi, interpreted in ranges. These included (i) Pa>Pi representing the potency of being active, (ii) Pa>0.7 denoting the high possibility of being experimentally active, (iii) 0.5<Pa<0.7 indicating the existing probability of being experimentally active, with substances distinct from established pharmaceutical agents, (iv) Pa<0.5 pointing to the low chance of result activity experimentally and a high new chemical entity [23-24].
Molecular docking
Protein preparation was started by downloading the crystallographic structure of the protein or enzyme from the Protein Data Bank (PDB) (https://www.rcsb.org). Each receptor protein was made by separating the original ligand using AutoDock Tool (ADT) 1.5.7 [25]. This tool was employed to verify and mend missing receptor atoms. Subsequently, the water molecules on the receptor protein were cleaned, added with Kollman charges, and saved in the PDBQT extension format. The separated original ligand was created by adding hydrogen atoms and Gasteiger charges. The processed protein and the original ligand were separated using specific parameters. The molecular docking of EGFR protein was carried out using a grid box with dimensions of 58×20×20 Å3 with Cartesian coordinates x, y, and z of 23.812, 0.303, and 52.113, respectively. Discovery Studio Visualizer (DSV) was used to eliminate the water molecules. Protein preparation was continued by adding hydrogen using AutoDock Tools 1.5.7 and saved in PDB format. Additionally, the ligand was initially prepared by sketching the hydrazone molecule's structure with Chemdraw Ultra 12.0 and keeping it in PDB format. The ligand in PDB format was entered into the AutoDock Tools 1.5.7 software for additional preparation. Rotatable bonds were adjusted in the torsion tree panel to account for the ligand's flexible characteristics in PDBQT format. Grid Box settings focused on ligands in protein structures, which were used to create configuration files. Docking simulation was carried out using AutoDock by entering commands, while the interactions between proteins and ligands were visualized using DSV.
ADMET-drug-likeness analysis
The drug-likeness and pharmacokinetic properties of benzylidene benzohydrazide derivatives compounds were analyzed using ADMETlab 2.0 (https://admetmesh.scbdd.com/service/evaluasi/index) [26]. The analysis method uses the chemical structure in SMILES format, which is input into ADMET LAB 2.0. Furthermore, ADMET LAB 2.0 will process, and the result is Drug-Likeness parameters such as Molecular Weight (MW), Log P (hydrophobicity), hydrogen bond acceptor (nHA), hydrogen bond donor (nHD), Total Polar Surface Area (TPSA). The pharmacokinetic variables evaluated included Human Intestinal Absorption (HIA) and Caco2 cells for uptake, Plasma Protein Binding (PPB) and Blood-Brain Barrier (BBB) for distribution, CYP inhibitors for metabolism, and Ames Toxicity for mutagenic and carcinogenic potential [27].
In vitro cytotoxic activity against CSC
The cytotoxicity of hydrazone ligands was examined with the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) tests [28]. 100 μl cell line was seeded into each well of a 96-well plate at a concentration of 5 x 103 cells/ml and adhered for 24 h in a CO2 incubator. Furthermore, this study used six replica wells as the control for comparison. Before treatment, the Cancer Stem Cells was grown for 24 h to achieve 80% confluency, and ten μL of the solution of the tested compounds was given to each well. The final concentrations of the tested compounds were maintained at 1.25, 2.5, 5, 10, 25, 50, and 100 μM. After 48 h of cultivation, 1 ml of MTT (50 mg of MTT/10 ml of PBS) was given to each well and then incubated for 4 h in a CO2 incubator. The formazan crystals produced were diluted in 100 μL of 10% SDS, placed in 0.01 N HCl, and the absorbance was examined with an ELISA reader at 595 nm wavelength. The SPSS statistic method was used to calculate the final IC50 values, and all tests were repeated at least three times in separate trials.
RESULTS
Anticancer profile prediction
Each benzylidene benzohydrazide derivative compound was predicted using the PASS server to observe its biological activity. Predictions with this server were made by identifying relevant biological activities and probability spectrums of the compounds. The analysis results are presented in table 1. The type of biological activity found that was relevant to the molecular analysis of these compounds was anticancer.
Table 1: Analysis of the anticancer activity of benzylidene benzo hydrazide derivatives using the PASS prediction server
| Compounds | Probabilities | Biological activity | |
| Pa | Pi | ||
| Erlotinib (native ligand) | 0.791 | 0.003 | Anticancer |
| Doxorubicin | 0.985 | 0.001 | Anticancer |
| Benzylidene benzohydrazide (L1) | 0.872 | 0.003 | Anticancer |
| 2-methylbenzylidene benzohydrazide (L2) | 0.825 | 0.004 | Anticancer |
| 2-nitrobenzylidene benzohydrazide (L3) | 0.769 | 0.004 | Anticancer |
| 2-bromobenzylidene benzohydrazide (L4) | 0.822 | 0.004 | Anticancer |
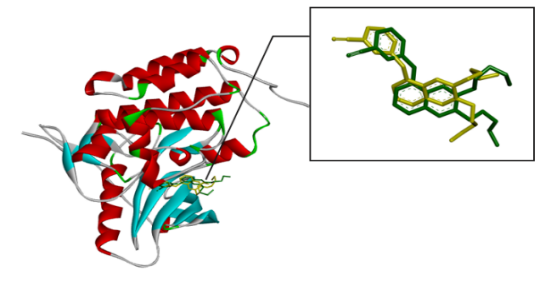
Fig. 1: Superimposed re-docking ligand (green) into native ligand (yellow) of protein receptor: the complex of EFGR (ID PDB: 1m17) – AQ4
Molecular docking
The beginning of docking procedures was the validation of the docking protocol with the EGFR receptor (PDB code: 1m17) and the native ligand erlotinib (AQ4). As shown in fig. 1, the native ligands were superimposed and included EGFR complexes with erlotinib (AQ4).
Table 2 exhibits the in silico docking test results between benzylidene benzohydrazide derivative hydrazone compounds and the EGFR receptor target (PDB code: 1m17). It reveals that the binding energy value of benzylidene benzohydrazide derivative compounds ranged from -6.69 to -7.74 kcal/mol. These values serve as indicators for predicting the activity of compounds. All benzylidene benzohydrazide derivative hydrazone compounds studied had lower binding energy values than their parent (binding energy =-6.69 kcal/mol).
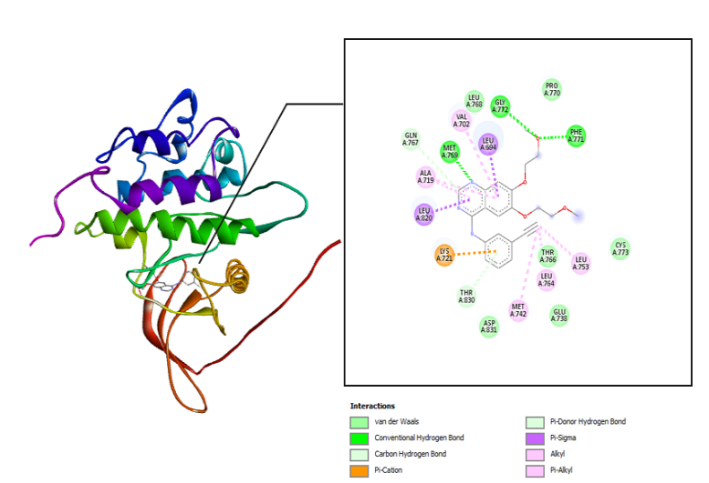
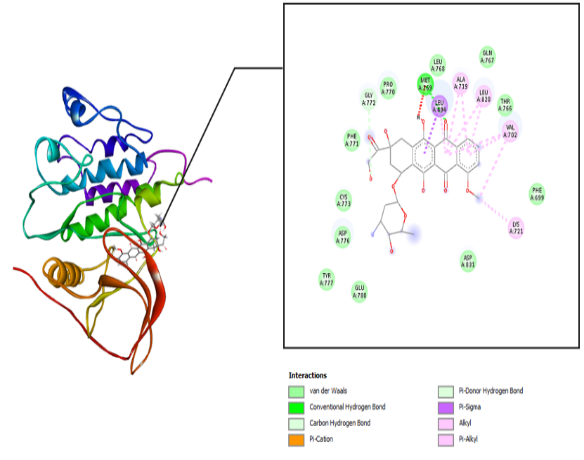
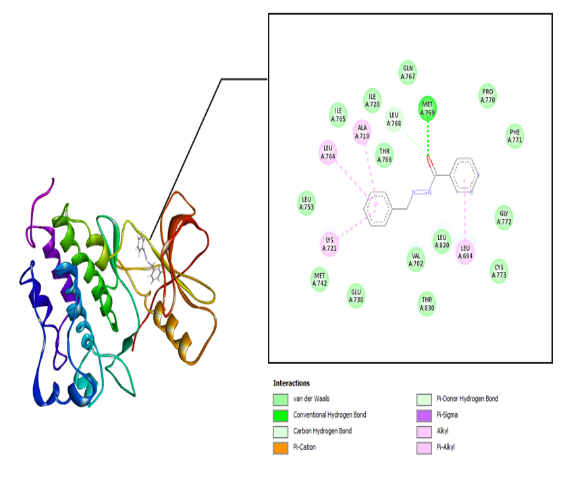
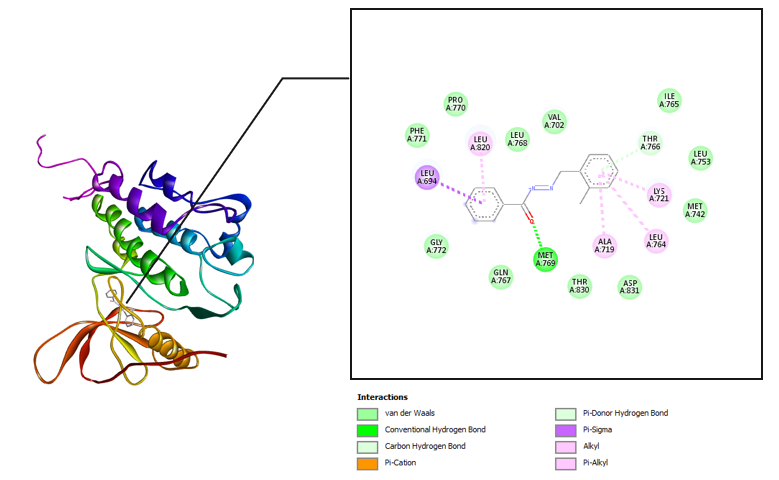
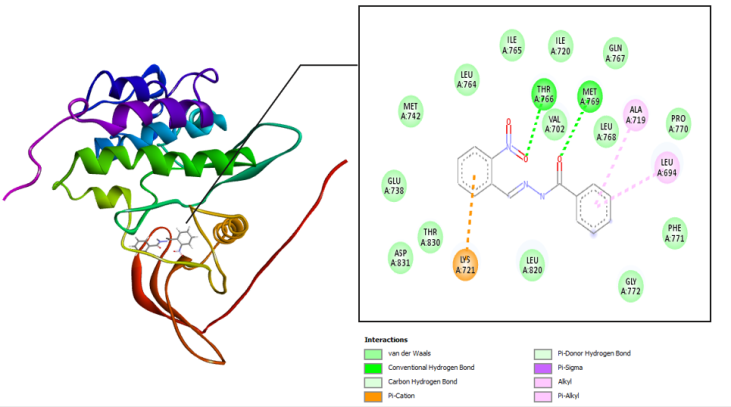
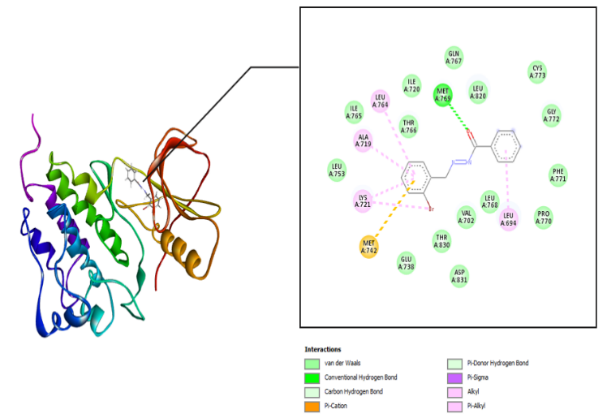
The interaction between the amino acid residues of the receptor and hydrazone ligand derivative benzylidene benzo hydrazide was formed through hydrogen bonds, Van der Waals, and hydrophobic interactions, as shown in table 3 and fig. 4-7. The similarity of amino acid residue between the original ligand and the test ligand showed the similarity of potential biological activity [29]. The binding conformational parameters showed the amino acid bonds formed by the ligand and receptor.
Table 2: Binding energy (Kcal/mol) of hydrazone derivative of benzaldehyde and benzo hydrazide
| Compounds | Chemical structure | Binding energy (Kcal/mol) |
| Erlotinib (native ligand) | 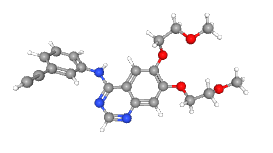 |
-8.64 |
| Doxorubicin |  |
-4.30 |
| Benzylidene benzohydrazide (L1) |  |
-6.69 |
| 2-methylbenzylidene benzohydrazide (L2) |  |
-6.70 |
2-nitrobenzylidene benzohydrazide (L3) |
 |
-7.34 |
| 2-bromobenzylidene benzohydrazide (L4) | 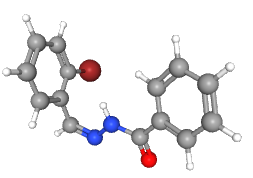 |
-7.44 |
Table 3: Result of molecular docking of EFGR with hydrazone derivative of benzaldehyde and benzo hydrazide
| Compounds | Amino acid residue interactions | |
| Hydrogen bond | Van der waals bond and hydrophobic interactions | |
| Erlotinib (native ligand) | MET769, GLY772, PHEA771 | THR766, LEU694, LEU820, VAL702, GLIN767, ALA719, LYS721, THR830, MET742, LEU753 |
| Doxorubicin | MET769 | GLY772, LEU694, ALA719, LEU820, VAL702, LYS721 |
| Benzylidene benzohydrazide (L1) | MET769 | LYS721, LEU764, ALA719, LEU768, LEU694 |
| 2-methylbenzylidene benzohydrazide (L2) | MET769 | LYS721, THR766, LEU764, ALA719, LEU694 |
| 2-nitrobenzylidene benzohydrazide (L3) | MET769, THR766 | LYS721, ALA719, LEU694 |
| 2-bromobenzylidene benzohydrazide (L4) | MET769 | ALA719, LEU764, LYS721, MET742, LEU694 |
(a) |
(b) |
Fig. 2: Molecular interaction: (a) the native ligand erlotinib and (b) doxorubicin with the EGFR receptor
(c) |
(d) |
Fig. 3: Molecular interactions: (c) benzylidene benzohydrazide and (d) 2-methyl benzylidene benzohydrazide with the EGFR receptor
(e) |
(f) |
Fig. 4: Molecular interactions: (e) 2-nitro benzylidene benzohydrazide and (f) 2-bromo benzylidene benzo hydrazide with the EGFR
Drug-likeness analysis
In silico predictions of parameter values for the physicochemical properties of benzylidene benzohydrazide derivative hydrazone compounds and reference compounds are shown in table 4. Lipinski's rules were used to evaluate the similarity of drugs to all compounds tested. Subsequently, a screening stage was done to determine the viability of the test compounds as drug candidates.
Table 4: Drug-likeness of hydrazone derivative of benzaldehyde and benzo hydrazide
| Compounds | MW | Log P | nHA | nHD | TPSA (Å) |
| Erlotinib (native ligand) | 393.170 | 2.497 | 7 | 1 | 77.960 |
| Doxorubicin | 543.170 | 1.760 | 12 | 9 | 212.390 |
| Benzylidene benzohydrazide (L1) | 224.090 | 3.634 | 3 | 1 | 41.460 |
| 2-methylbenzylidene benzohydrazide (L2) | 238.110 | 3.964 | 3 | 1 | 41.460 |
| 2-nitrobenzylidene benzohydrazide (L3) | 269.080 | 3.445 | 6 | 1 | 84.600 |
| 2-bromobenzylidene benzohydrazide (L4) | 302.010 | 4.196 | 3 | 1 | 41.460 |
Prediction of absorption, distribution, metabolism, excretion and toxicity (ADMET)
The results of the ADMET prediction for hydrazone derivatives based on benzaldehyde and benzohydrazide compounds are shown in table 5.
Table 6 exhibits the metabolic profile of a drug can be assessed by evaluating the inhibitory effect of compounds on cytochrome enzymes. Cytochrome P450 (CYP) enzymes are a superfamily of isoenzymes, serving a crucial function in drug removal via metabolic biotransformation [30]. CYP450 generally has five primary isoforms, including CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4.
Table 5: ADMET prediction of hydrazone derivative and reference compounds using ADMETLab 2.0
| Compounds | Absorption | Distribution | Excretion | Toxicity | QED score | |||
| HIA (%) | Caco-2 (10-6 cm/s) | PPB (%) | BBB | (CL) | AMES toxicity | Carcinogenicity | ||
| Erlotinib | ≥ 30 | -4.668 | 91.222 | Excellent | High | Medium | Excellent | 0.447 |
| Doxorubicin | ≤ 30 | -6.006 | 91.288 | Excellent | High | Poor | Poor | 0.147 |
| Benzylidene benzohydrazide (L1) | ≥ 30 | -4.466 | 98.278 | Medium | Poor | Excellent | Medium | 0.631 |
| 2-methylbenzylidene benzohydrazide (L2) | ≥ 30 | -4.466 | 98.278 | Excellent | High | Excellent | Medium | 0.648 |
| 2-nitrobenzylidene benzohydrazide (L3) | ≥ 30 | -4.461 | 98.316 | Excellent | Poor | Poor | Poor | 0.525 |
| 2-bromobenzylidene benzohydrazide (L4) | ≥ 30 | -4.423 | 98.785 | Medium | Poor | Excellent | Medium | 0.686 |
Table 6: Metabolism prediction profile of hydrazone derivative compounds and reference compounds using ADMETLab 2.0
| Compounds | Cytochrome P450 (CYP) enzyme inhibitors | ||||
| 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | |
| Erlotinib | Inhibitor | Inhibitor | Inhibitor | Inhibitor | Inhibitor |
| Doxorubicin | No | No | No | No | No |
| Benzylidene benzohydrazide | Inhibitor | No | No | No | No |
| 2-methylbenzylidene benzohydrazide | Inhibitor | Inhibitor | Inhibitor | No | No |
| 2-nitrobenzylidene benzohydrazide | Inhibitor | Inhibitor | Inhibitor | No | No |
| 2-bromobenzylidene benzohydrazide | Inhibitor | Inhibitor | Inhibitor | No | No |
In vitro cytotoxic activity against CSCs
The MTT method performed (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) with doxorubicin positive control to assess the cytotoxic activity of hydrazone ligand substances tested on CSCs. Fig. 5 shows the microscopic morphology of Cancer Stem Cells (CSCs) after treatment with hydrazone compounds.
Note: () indicates normal living cells
The morphology of cancer stem cells after undergoing apoptosis and necrosis is shown in fig. 6.
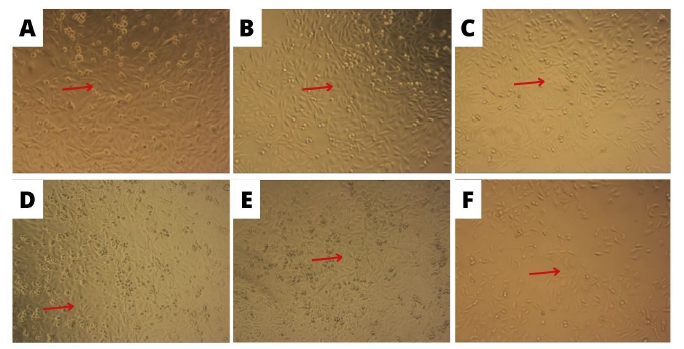
Fig. 5: Morphology of cancer stem cells treated with test compounds with incubation for 24 h with 10x magnification (A) control cells (B) benzylidene benzo hydrazide (C) 2-methyl benzylidene benzohydrazide (D) 2-bromobenzylidene benzo hydrazide (E) 2-nitro benzylidene benzohydrazide (F) doxorubicin
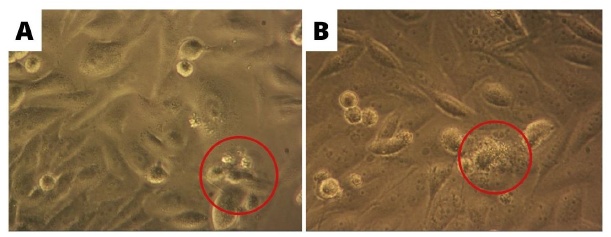
Fig. 6: Cancer Stem Cells that morphological changes after being treated with the test compound were observed with an inverted microscope at 40x magnification (A) apoptosis (B) necrosis
Table 7 shows the in vitro cytotoxicity evaluation of the hydrazone compounds.
Table 7: The in vitro cytotoxicity evaluation of hydrazone compounds
| Sample | CSC |
| IC50 (μg/ml) | |
| Doxorubicin | 0.220±0.180 |
| Benzylidene benzohydrazide | 0.882±0.360 |
| 2-methylbenzylidene benzohydrazide | 0.034±0.023 |
| 2-nitrobenzylidene benzohydrazide | 0.355±0.276 |
| 2-bromobenzylidene benzohydrazide | 1.193±1.122 |
DISCUSSION
The result of the analysis from the Prediction of Activity Spectra for Substances (PASS) server showed that all compounds had a probability of being active as anticancer based on the active probability value (Pa) being more significant than the inactive probability value (Pi). The hydrazone derivative analyzed showed Pa values between 0.769 and 0.872. Meanwhile, the highest Pa value was shown by the compounds Benzylidene benzohydrazide, followed by 2-methylbenzylidene benzohydrazide, 2-bromobenzylidene benzohydrazide, and 2-bromobenzylidene benzohydrazide. The prediction results show that all hydrazone compounds derived from benzohydrazide and benzaldehyde have potential anticancer properties. Furthermore, the interaction of these compounds can be confirmed through molecular docking.
The molecular docking was conducted to determine the interactions that occurred between erlotinib, doxorubicin, benzylidene benzohydrazide, 2-methylbenzylidene benzohydrazide, 2-nitrobenzylidene benzohydrazide, and 2-bromobenzylidene benzohydrazide on the EGFR receptor on CSCs (PDB code: 1m17). As shown in fig. 1, the native ligands were superimposed and included EGFR complexes with erlotinib (AQ4). The validation results provided a root-mean-square deviation (RMSD) value of 1.42 Å, a vital parameter to forecast the possible activity of substances, which is less than 2Å [31]. Furthermore, all hydrazone compounds studied had binding energy values more significant than the native ligand erlotinib (binding energy =-8.64 kcal/mol) but smaller compared to doxorubicin (binding energy =-4.30 Kcal/mol). As shown in table 2, the binding energies for 2-nitro benzylidene benzohydrazide and 2-methyl benzylidene benzohydrazide are-7.34 Kcal/mol and 6.70 Kcal/mol, respectively. The 2-bromobenzylidene benzohydrazide compounds had the smallest binding energy, namely-7.44 Kcal/mol. This showed that 2-bromobenzylidene benzohydrazide had the best interaction in binding to the EGFR receptor amino acids, making it more stable. Binding energy refers to the energy of intermolecular interactions between receptors and ligands. A lower binding energy means that the interaction of ligand and receptor has more potential to occur, indicating excellent activity [32, 33]. The binding conformational parameters showed the amino acid bonds formed by the ligand and receptor. The low binding energy value of the 2-bromobenzylidene benzohydrazide compounds was attributed to the hydrogen bond between the oxygen in the carbonyl group and the amino acid MET769, the pi-alkyl interaction between the benzene ring and the amino acids LEU694, LEU764, ALA719, and LYS721, as well as the pi-sulfur interaction between the benzene rings and amino acid MET742.
The results of the drug-likeness analysis showed that the test and reference compounds would be easily absorbed, with high permeability due to their optimal molecular weight ranging from 100 to 600 g/mol. All test compounds' log octanol/water partition coefficient (log P) values were non-optimal, falling outside the 0 – 3 log mol/l range. The donor H-bond (nHD), represented by the count of O-H and N-H groups for all tested compounds, exhibited an optimal nHD value from 0 to 7. Furthermore, the acceptor H-bond (nHA) value showed the number of O and N atoms. All test compounds had optimal nHA values as drug candidates ranging from 0 to 12. The test compounds' Topological Polar Surface Area (TPSA) values were optimal, as shown by 0 – 140 Å. Table 4 shows that all test compounds fulfilled the Lipinski rule. Lipinski's law is considered for active substances given orally, and its criteria are the first step in assessing the oral bioavailability of active elements pertaining to the solubility and permeability of substances in the gastrointestinal tract [34, 35].
Based on the results of the ADMET prediction for hydrazone derivative compounds, the Quantitative Estimate of Druglikeness (QED) value ranged from 0.147 to 0.686. The QED Score predicts the similarity of a chemical compound's property to establish drugs, using parameters such as molecular mass, lipophilicity, and polarity, including several relevant physical and chemical factors. All compounds studied had QED values greater than the native ligand erlotinib (QED = 0.447) and the reference compounds doxorubicin (QED = 0.147). Compounds 2-bromobenzylidene benzohydrazide had the largest QED value, namely 0.686, making it relevant as an effective drug. The HIA value shows the absorption level of active substances in the human digestive tract. Generally, compounds are appropriately absorbed when the % HIA value is in the range ˃ 30%. All test compounds and native ligands had good HIA values, but doxorubicin was in the<30% range, while high Caco-2 cell permeability was>-5.15. All test compounds had Caco-2 cell values<-5.15, showing low permeability in penetrating cell membranes. Meanwhile, doxorubicin had a high permeability value, namely 6.006. PPB percentages affect the pharmacokinetic and pharmacodynamic characteristics of drugs. In this study, all test compounds had PPB values ≥ 90%, showing that the drug was firmly bound to plasma proteins, making distribution to the target of action difficult. Another distribution parameter is the BBB, which shows the concentration of a drug in the brain. This parameter is essential for assessing the drug's capability in penetrating the BBB. According to the analysis results using ADMETLab 2.0, all compounds, both test compounds and native ligands, showed good BBB. The highest Clearance of Drug (CL) value was found in compounds 2-methylbenzylidene benzohydrazide, showing that compounds could be excreted well. The AMES test observed the toxicity profile through mutagenicity and carcinogenicity parameters. Meanwhile, the AMES test assessed the toxicity profile through mutagenicity and carcinogenicity parameters [36]. Almost all test compounds were not mutagens or carcinogens.
The results of the metabolism prediction profile of hydrazone derivative compounds are shown in table 6. Inhibiting this isoenzyme leads to drug interactions related to pharmacokinetics, resulting in harmful side impacts or adverse drug reactions due to decreased clearance and accumulation [37]. Based on table 6, it was discovered that all test compounds were predicted to have the potential to inhibit three main isoforms, namely 1A2, 2C19, and 2C9 of the cytochrome P450 enzyme.
The in vitro cytotoxic activity test results against CSCs are shown in fig 5-6 and table 7. The principle of the process included reducing the MTT solution to formazan salt by involving the enzyme succinate dehydrogenase found in the mitochondria of living cells [38]. Based on the observation data, the microscopic morphology of Cancer Stem Cells (CSCs) is presented in fig. 5. Cancer Stem Cells exposed to the test compound for 24 h appeared to change their shape compared to cells without treatment (control cells). Cell death caused by osmotic shock or differences in fluid concentration inside or outside the cell causes the cell to lose the integrity of its cell membrane, followed by an enlarged nucleus, which includes uncontrolled cell death or necrosis and can cause inflammatory effects [39].
The intercalation and free radicals by the compounds cause DNA in the nucleus to lose its transcription ability and the replication system, thereby affecting other cellular processes [40]. DNA damage will then cause cells to undergo apoptosis [41]. Cells treated with the test compound showed morphological changes that led to the characteristics of cells undergoing apoptosis, namely shrinkage and cytoplasm compaction and damage to the extracellular matrix. In fig. 6, cells can be seen undergoing apoptosis and necrosis. Changes in cell morphology due to exposure to certain active compounds or chemotherapy agents reflect biochemical conditions, causing cell death either by apoptosis or necrosis.
The results of the cytotoxicity test of test compounds with CSCs provide IC50 results for L1 of 0.220±0.360 μg/ml, L2 of 0.034±0.023 μg/ml, L3 of 0.355±0.276 μg/ml, and L4 of 1.193±1.122 μg/ml. All benzylidene benzohydrazide derivatives have very active cytotoxic activity (IC50<1 µg/ml) [42–44]. The various results of the cytotoxicity evaluation of the four test compounds were due to the addition of different groups to each compound. Doxorubicin showed IC50 values below one µg/ml (0.220±0.180 μg/ml). The IC50 values of test compounds were comparable and even lower than doxorubicin, so they can be considered new anticancer therapeutic candidates.
CONCLUSION
In silico studies show that the compound 2-bromobenzylidene benzohydrazide has a lower binding energy (-7.44 kcal/mol) compared to other compounds, making it more stable in forming bonds with receptors. Based on the results of in vitro tests on CSCs, it was shown that the compound 2-methylbenzylidene benzohydrazide has a minimal IC50 value, namely 0.034 μg/ml, meaning that 2-methylbenzylidene benzohydrazide has very strong cytotoxicity compared to other test compounds. In silico and in vitro results also showed that compound 2-bromobenzylidene benzo hydrazide and 2-methylbenzylidene benzohydrazide had the potential as a CSCs inhibitor.
ACKNOWLEDGMENT
The authors thank the Rector of Sepuluh Nopember Institute of Technology (ITS) for the financial support through Scientific Research Grants (2216/PKS/ITS/2023).
AUTHORS CONTRIBUTIONS
Conceptualization, I. G., Y. S. A and F. M.; methodology, I. G., Y. S. A., and M. S.; software, V. A. N. A and N. R. A.; validation, M. S., A. F. and Z. Z.; formal analysis, F. M.; investigation, P. P.; resources, I. G and F. M.; data curation, Y. S. A.; writing—original draft preparation, I. G.; writing—review and editing, P. P and F. M.; visualization, A. F.; supervision, M. S.; project administration, N. R. A.; funding acquisition, F. M. All authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTERESTS
There are no conflicts of interest to disclose in this research.
REFERENCES
Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124-34. doi: 10.1038/nm.4409, PMID 28985214.
Das M, Law S. Role of tumor microenvironment in cancer stem cell chemoresistance and recurrence. Int J Biochem Cell Biol. 2018 Oct;103:115-24. doi: 10.1016/j.biocel.2018.08.011, PMID 30153480.
Zhang C, Yang Z, Dong DL, Jang TS, Knowles JC, Kim HW. 3D culture technologies of cancer stem cells: promising ex vivo tumor models. J Tissue Eng. 2020;11:2041731420933407. doi: 10.1177/2041731420933407, PMID 32637062.
Liu Y, Zhang Y, Chen S, Zhong X, Liu Q. Effect of LGR4/EGFR signaling on cell growth and cancer stem cell-like characteristics in liver cancer. Cytokine. 2023 May;165:156185. doi: 10.1016/j.cyto.2023.156185, PMID 37001327.
Nasrolahi A, Azizidoost S, Radoszkiewicz K, Najafi S, Ghaedrahmati F, Anbiyaee O. Signaling pathways governing glioma cancer stem cells behavior. Cell Signal. 2023 Jan;101:110493. doi: 10.1016/j.cellsig.2022.110493, PMID 36228964.
Talukdar S, Emdad L, Das SK, Fisher PB, Chapter Four EG. An essential receptor tyrosine kinase regulator of cancer stem cells. In: Receptor tyrosine kinases. Kumar R, Fisher PB, editors. Academic Press; 2020. p. 161-88. Available from: https://www.sciencedirect.com/science/article/pii/S0065230X2030035X.
Xie X, Qiu G, Chen Z, Liu T, Yang Y, You Z. Characteristics and prognosis of EGFR mutations in small cell lung cancer patients in the NGS era. Clin Transl Oncol. 2024;26(2):434-45. doi: 10.1007/s12094-023-03263-w, PMID 37436674.
XU G, Abad MC, Connolly PJ, Neeper MP, Struble GT, Springer BA. 4-amino-6-arylamino pyrimidine-5-carbaldehyde hydrazones as potent ErbB-2/EGFR dual kinase inhibitors. Bioorg Med Chem Lett. 2008;18(16):4615-9. doi: 10.1016/j.bmcl.2008.07.020, PMID 18653333.
Duan H, Liu Y, Gao Z, Huang W. Recent advances in drug delivery systems for targeting cancer stem cells. Acta Pharm Sin B. 2021;11(1):55-70. doi: 10.1016/j.apsb.2020.09.016, PMID 33532180.
Sun TM, Wang YC, Wang F, DU JZ, Mao CQ, Sun CY. Cancer stem cell therapy using doxorubicin conjugated to gold nanoparticles via hydrazone bonds. Biomaterials. 2014;35(2):836-45. doi: 10.1016/j.biomaterials.2013.10.011, PMID 24144908.
Alotabi SH. Synthesis characterization anticancer activity and molecular docking of some new sugar hydrazone and arylidene derivatives. Arab J Chem. 2020;13(3):4771-84. doi: 10.1016/j.arabjc.2019.12.006.
Sharma JD, Dutta SKDKJPP, Kumar D. From tetralone: synthesis spectral characterization. In Vitro. 2021.
Aly SA, Fathalla SK. Preparation characterization of some transition metal complexes of hydrazone derivatives and their antibacterial and antioxidant activities. Arab J Chem. 2020;13(2):3735-50. doi: 10.1016/j.arabjc.2019.12.003.
Verma G, Marella A, Shaquiquzzaman M, Akhtar M, Ali MR, Alam MM. A review exploring biological activities of hydrazones. J Pharm Bioallied Sci. 2014;6(2):69-80. doi: 10.4103/0975-7406.129170, PMID 24741273.
Arjun HA, Elancheran R, Manikandan N, Lakshmithendral K, Ramanathan M, Bhattacharjee A. Design synthesis and biological evaluation of (E)-N-((1-chloro-3,4-Dihydronaphthalen-2-yl)methylene)benzohydrazide derivatives as antiprostate cancer agents. Front Chem. 2019 Jul 10;7. doi: 10.3389/fchem.2019.00474.
MO QY, Deng JG, Liu Y, Huang GD, LI ZW, YU P. Mixed ligand Cu(II) hydrazone complexes designed to enhance anticancer activity. Eur J Med Chem. 2018 Aug 5;156:368-80. doi: 10.1016/j.ejmech.2018.07.022, PMID 30015073.
Wermuth CG, Ganellin CR, Lindberg P, Mitscher LA. Glossary of terms used in medicinal chemistry (IUPAC Recommendations 1998). Pure Appl Chem. 1998;70(5):1129-43. doi: 10.1351/pac199870051129.
Frimayanti N, Mora E, Anugrah R. Study of molecular docking of chalcone analoque compound as inhibitors for liver cancer cells HepG2. Com Eng App. 2018;7(2):137-47. doi: 10.18495/comengapp.v7i2.260.
Frimayanti N, Ikhtiarudin I, Dona R, Agustini TT, Murdiya F, Zamri A. A computational approach to drug discovery: search for chalcone analogues as the potential candidates for anti colorectal cancer (HT29). Walailak J Sci Technol. 2018;17(2):64-74. doi: 10.48048/wjst.2020.5910.
Lesage R, Blanco MN, Narcisi R, Welting T, Van Osch GJ, Geris L. An integrated in silico in vitro approach for identification of therapeutic drug targets for osteoarthritis. Bio Rxiv; 2021. Available from: https://www.biorxiv.org/content/early/2021/09/27/2021.09.27.461207.
Alam YS, Pudjiastuti P, Maulana S, Afifah NR, Martak F, Fadlan A. Synthesis and antidiabetic evaluation of N-benzylidenebenzohydrazide derivatives by in silico studies. Indones J Chem. 2023;23(4):1061-70. doi: 10.22146/ijc.82073.
Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277(48):46265-72. doi: 10.1074/jbc.M207135200, PMID 12196540.
Abdelli I, Hassani F, Bekkel Brikci S, Ghalem S. In silico study the inhibition of angiotensin-converting enzyme 2 receptor of COVID-19 by ammoides verticillata components harvested from Western Algeria. J Biomol Struct Dyn. 2021;39(9):3263-76. doi: 10.1080/07391102.2020.1763199, PMID 32362217.
Shah FH, Lim KH, Kim SJ. Do fever relieving medicines have anti-COVID activity: an in silico insight. Future Virol. 2021;16(4):293-300. doi: 10.2217/fvl-2020-0398.
Kawsar SM, Hossain MA, Hosen MI, Parmar MP, Patel SG, Patel HM. Novel benzylidene derivatives: synthesis and their antimicrobial and anticancer studies and in silico investigations. Chem Phys Impact. 2025 Jun;10:100786. doi: 10.1016/j.chphi.2024.100786.
Xiong G, WU Z, YI J, FU L, Yang Z, Hsieh C. ADMET lab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49(W1):W5-W14. doi: 10.1093/nar/gkab255, PMID 33893803.
Gao J, Liu X, Zhang B, Mao Q, Zhang Z, Zou Q. Design synthesis and biological evaluation of 1-alkyl-5/6-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)-1H-indole-3-carbonitriles as novel xanthine oxidase inhibitors. Eur J Med Chem. 2020;190:112077. doi: 10.1016/j.ejmech.2020.112077, PMID 32014678.
Cholayil Palapetta S, Gurusamy H, Ganapasam S. Synthesis characterization computational studies molecular docking and in vitro anticancer activity of dihydropyrano[3,2-c]chromene and 2-aminobenzochromene derivatives. ACS Omega. 2023;8(8):7415-29. doi: 10.1021/acsomega.2c06049, PMID 36873031.
Kelutur FJ, Mustarichie R. Molecular docking of the potential compound from cocoa shells (Theobroma cacao l.) against androgen receptor as anti-alopecia. J Glob Pharm Technol. 2020;12(9):52-60.
Kirchmair J, Goller AH, Lang D, Kunze J, Testa B, Wilson ID. Predicting drug metabolism: experiment and/or computation? Nat Rev Drug Discov. 2015;14(6):387-404. doi: 10.1038/nrd4581, PMID 25907346.
Hastuti LP, Hermawan F, Iresha MR, Ernawati T. In silico studies of hydroxyxanthone derivatives as potential pfDHFR and pfDHODH inhibitors by molecular docking molecular dynamics simulation MM-PBSA calculation and pharmacokinetics prediction. Informatics Med Unlocked. 2024 Nov;47:101485. doi: 10.1016/j.imu.2024.101485.
Rifai EA, Van Dijk M, Vermeulen NP, Yanuar A, Geerke DP. A comparative linear interaction energy and MM/PBSA study on SIRT1 ligand binding free energy calculation. J Chem Inf Model. 2019;59(9):4018-33. doi: 10.1021/acs.jcim.9b00609, PMID 31461271.
De Lima Menezes G, Sales Bezerra K, Nobre Oliveira JI, Fontenele Araujo J, Soares Galvao D, Alves Da Silva R. Quantum mechanics insights into melatonin and analogs binding to melatonin MT1 and MT2 receptors. Sci Rep. 2024;14(1):10922. doi: 10.1038/s41598-024-59786-x, PMID 38740789.
SS, Smith, AA VV. Computational molecular modelling of 2-aminobenzimidazole derivatives: strong successor of hypoglycaemic agent. Int J Pharm Pharm Sci. 2021;13(9):16-21. doi: 10.22159/ijpps.2021v13i9.41877.
Mathew C, Lal N, SL TRA, Varkey J. Antioxidant anticancer and molecular docking studies of novel 5-benzylidene substituted rhodanine derivatives. Int J Pharm Pharm Sci. 2023;15(7):7-19. doi: 10.22159/ijpps.2023v15i7.47421.
Praveen M. Evaluation of antagonist activity of ifenprodil and their analogous against Glun1/Glun2B using in silico molecular docking and absorption distribution metabolism excretion toxicity. Asian J Pharm Clin Res. 2022;15(5):34-40. doi: 10.22159/ajpcr.2022.v15i5.44248.
Agus AS, Siswandono BB, Taufiqurrahman M, Fernandes A, Maharani R. Molecular docking of the keruings (Dipterocarpus) genus secondary metabolites of the Dipterocarpaceae family’s as anti-inflammation against cyclooxygenase-2 (COX-2). Int J Appl Pharm. 2024;16(2):313-9.
Das J, Debbarma A, Lalhlenmawia H. Formulation and in vitro evaluation of poly-(D, L-Lactide-Co-Glycolide) (Plga) nanoparticles of ellagic acid and its effect on human breast cancer Mcf-7 cell line. Int J Curr Pharm Sci. 2021;13(5):56-62. doi: 10.22159/ijcpr.2021v13i5.1887.
Zhang G, Wang J, Zhao Z, Xin T, Fan X, Shen Q. Regulated necrosis a proinflammatory cell death potentially counteracts pathogenic infections. Cell Death Dis. 2022;13(7):637. doi: 10.1038/s41419-022-05066-3, PMID 35869043.
Shakeel M, Butt TM, Zubair M, Siddiqi HM, Janjua NK, Akhter Z. Electrochemical investigations of DNA intercalation potency of bisnitrophenoxy compounds with different alkyl chain lengths. Heliyon. 2020;6(6):e04124. doi: 10.1016/j.heliyon.2020.e04124, PMID 32548325.
Toyoshima Sasatani M, Imura F, Hamatake Y, Fukunaga A, Negishi T. Mutation and apoptosis are well coordinated for protecting against DNA damage-inducing toxicity in drosophila. Genes Environ. 2023;45(1):11. doi: 10.1186/s41021-023-00267-4, PMID 36949493.
Jablonska E, Kubasek J, Vojtech D, Ruml T, Lipov J. Test conditions can significantly affect the results of in vitro cytotoxicity testing of degradable metallic biomaterials. Sci Rep. 2021;11(1):6628. doi: 10.1038/s41598-021-85019-6, PMID 33758226.
Zaki AA, Al Karmalawy AA, Khodir AE, El Amier YA, Ashour A. Isolation of cytotoxic active compounds from reichardia tingitana with investigation of apoptosis mechanistic induction: in silico in vitro and SAR studies. S Afr J Bot. 2022 Jan;144:115-23. doi: 10.1016/j.sajb.2021.08.006.
Febriansah R, Hertiani T, Widada J, Taher M, Damayanti E, Mustofa M. Isolation of active compounds from streptomyces sennicomposti GMY01 and cytotoxic activity on breast cancer cells line. Heliyon. 2024;10(2):e24195. doi: 10.1016/j.heliyon.2024.e24195, PMID 38293453.