
Int J App Pharm, Vol 17, Issue 5, 2025, 263-270Original Article
DEVELOPMENT OF REDUCED GRAPHENE OXIDE (RGO) NANOSHEETS FOR ULTRASENSITIVE DETECTION OF A NOVEL CORONA VIRUS SARS-COV-2
DEEPAK V. SAWANTa*, PRANAV K. KATKARb, ARUNKUMAR PARTHASARATHYa, R. K. SHARMAa, SHIVAJI KASHTEc, C. D. LOKHANDEd
aDepartment of Microbiology, Dr. D. Y. Patil Medical College Hospital and Research Institute Kolhapur-416006, Maharashtra, India. bDepartment of Physics, Sejong University, Gwangjin-gu, Seoul-05006, Republic of Korea. c,dCentre for Interdisciplinary Research, D. Y. Patil Education Society (Deemed to be University), Kolhapur-416006, Maharashtra, India
*Corresponding author: Deepak V. Sawant; *Email: sawantlab@gmail.com
Received: 09 Dec 2024, Revised and Accepted: 17 Jun 2025
ABSTRACT
Objective: The extraction of ribonucleic acid (RNA) is essential for SARS-CoV-2 RT-PCR detection. Although commercial RNA extraction kits are available, they suffer from limitations such as low RNA yield, difficulty in amplification during RT-PCR, and high costs. This study investigates the evolution of rGO Nano sheets as a novel material for efficient RNA extraction from clinical specimens. The rGO Nano sheets offer a promising alternative for the purification of excellent RNA, which is crucial for accurate and high-sensitivity identification of SARS-CoV-2. By leveraging a unique property of rGO, the research demonstrates enhanced RNA recovery, improving the overall sensitivity and reliability of detection method for SARS-CoV-2.
Methods: The rGO Nano sheets were synthesized by reducing graphene oxide (GO) using a simple hydrothermal process. The rGO analysis and its interaction with RNA were characterized using X-ray diffraction, Raman spectroscopy, field emission scanning electron microscopy (FE-SEM), and Brunauer-Emmett-Teller (BET) analysis. RNA was extracted from clinical specimens using rGO Nano sheets via adsorption and elution and compared with extraction performed using a commercial RNA kit. RNA extraction efficiency and validation were assessed by UV-visible spectrometry and RT-PCR.
Results: The rGO Nano sheets exhibited a multi-layered carbon nanostructure with an average 7.15 nm 3D porous structure, a large surface area of 225 m²/g, and interconnected carbon Nano sheets. The RNA extracted using rGO Nano sheets was validated by UV-visible spectrometry and RT-PCR, showing successful detection of viral genes (ORF1ab and N gene) with a sensitivity of 10 copies. Ten to twenty copies of SARS-CoV-2 pseudo-virus particles showed a strong linear association. The P-value is greater than 0.05, indicating acceptance of the null hypothesis. The commercial kit and the rGO Nano sheet-based RNA extraction methods are not significantly different from one another. There is a significant increase in RNA yield (1.959 ng/µl) and improved sensitivity in RT-PCR, further validating the advantage of using rGO for RNA extraction.
Conclusion: The developed rGO Nano sheet-based RNA extraction method demonstrates high sensitivity and efficiency, offering a promising alternative to commercial kits for the detection of SARS-CoV-2.
Keywords: Reduced graphene oxide, Ribonucleic acid, SARS-CoV-2, COVID-19 detection, Nano-sheets
© 2025 The Authors. Published by Innovare Academic Sciences Pvt Ltd. This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/)
DOI: https://dx.doi.org/10.22159/ijap.2025v17i5.53365 Journal homepage: https://innovareacademics.in/journals/index.php/ijap
INTRODUCTION
COVID-19, caused by the SARS-CoV-2 virus, was first identified in December 2019 and has since led to a global pandemic, with over 614 million cases and 6.5 million deaths as of October 2022 [1, 2]. SARS-CoV-2 is an enveloped virus with a positive-sense, single-stranded RNA genome. Since its initial discovery, COVID-19 has spread globally, resulting in a pandemic with significant morbidity and mortality [1, 2]. SARS-CoV-2 exhibits significant genetic variability due to frequent mutations and recombination [3, 4]. Accurate and timely diagnosis of COVID-19 is critical, and RT-PCR has become the gold standard for detection. However, extracting high-quality viral RNA from patient samples remains a significant challenge. The process typically involves viral inactivation using detergents [5,6], followed by RNase denaturation with guanidinium salts [7] or proteases like proteinase K. Contaminants from these reagents, such as residual guanidinium salts or organic solvents, can interfere with RT-PCR, necessitating further purification. Current RNA extraction methods, such as Trizol-based liquid separation or solid-phase silica extraction [5], often suffer from low yield and purity. To address these limitations, reduced graphene oxide (rGO) Nano sheets have shown promise. Their positively charged surface enables efficient RNA binding, especially in the presence of guanidinium salts and alkaline lysis buffers [8]. This results in higher RNA yield and improved purity, making rGO Nano sheets a promising alternative for enhancing RNA extraction from clinical samples.
MATERIALS AND METHODS
Sigma-Aldrich supplied the sodium nitrate (NaNO₃) and graphite flakes, sulphuric acid (H₂SO₄), potassium permanganate (KMnO₄), carbon black, N-Methyl-2-Pyrrolidone (NMP), and polyvinylidene fluoride (PVDF). Additionally, Alfa-Aesar (India) supplied the potassium hydroxide (KOH) and hydrogen peroxide (H₂O₂). All chemicals used were of AR grade and did not require any additional purification.
Synthesis of reduced graphene oxide (RGO) nano sheets
Reduced graphene oxide (rGO) Nano sheets were synthesized using a modified Hummers’ method followed by thermal reduction. Briefly, graphite powder was oxidized using a combination of strong sodium nitrate (NaNO₃), potassium permanganate (KMnO₄), and sulphuric acid (H₂SO₄), to produce graphene oxide (GO). The GO was then reduced thermally at 800 °C under an inert atmosphere to obtain rGO Nano sheets.
Characterization of rGO nano sheets
The synthesized rGO Nano sheets were characterized using the following techniques to confirm their structural, morphological, and functional specifications XRD analyzed the phase and crystal transparency of rGO. The disappearance of the GO peaks and the appearance of a broad rGO peak confirmed successful reduction. Measured the defect density and structural integrity of rGO. The intensity ratio of the D band (defects) to the G band (graphitic structure) provided insights into the reduction efficiency. Transmission Electron Microscopy (TEM): Visualized the morphology and layer structure of rGO Nano sheets, confirming their exfoliation and nanoscale dimensions. Fourier-Transform Infrared Spectroscopy (FTIR): Identified functional clusters on the rGO surface, ensuring the elimination of oxygen-containing clusters after reduction.
RNA Extraction using rgo nano sheets
Viral RNA extraction from SARS-CoV-2 clinical samples was performed using rGO Nano sheets. Sample Preparation: Nasopharyngeal swab samples were collected and suspended in guanidinium thiocyanate and Triton X-100 lysis buffer to inactivate the virus and release RNA. The lysis buffer with rGO Nano sheets was incubated at room temperature for 15 min with RNA binding process. The positively charged surface of rGO facilitated the adsorption of negatively charged RNA. The rGO-RNA complex was washed twice with 70% ethanol to remove impurities such as proteins and salts. Elution: RNA was eluted from the rGO Nano sheets using nuclease-free water at pH 8.0.
Controls for RNA extraction
A positive control (a known concentration of SARS-CoV-2 RNA) was processed alongside biological specimens to ensure the efficiency of RNA binding and elution. A negative control (nuclease-free water without RNA) was used to confirm the absence of contamination. A commercial RNA extraction kit available in the market was used as a benchmark to compare the performance of the rGO-based method.
RNA quantification and purity assessment
The concentration and purity of extracted RNA were quantified using UV-Vis spectroscopy. RNA samples were diluted in nuclease-free water, and absorbance was measured at 260 nm (A260) and 280 nm (A280) using a spectrophotometer. UV-Vis Spectroscopy: UV-Vis spectroscopy is a widely used technique for nucleic acid quantification due to its simplicity, speed, and reliability. The spectral absorption at 260 nm (A260) is proportional to RNA absorption, while the A260/A280 ratio provides an estimate of RNA purity. A ratio of ~2.0 indicates high-purity RNA, free from protein contamination. To ensure accurate quantification a typical arch was generated with well-known absorptions of RNA. Each sample was measured in triplicate to account for variability. Background correction was performed using a blank (nuclease-free water) to eliminate interference from the solvent.
Validation of extracted RNA
The quality and integrity of extracted RNA were further validated with RT-PCR, isolated RNA was reverse transcribed into complementary DNA (cDNA) followed by amplification of the E-gene and RdRp-gene of SARS-CoV-2 to confirm the presence of viral RNA. Agarose Gel electrophoresis RNA samples were run on a 1.5% agarose gel to visualize intact RNA bands and assess degradation.
Statistical examination
Data were presented as mean±standard deviation (SD), and all experiments were run in triplicate. Student's t-test was used to establish statistical significance, with p<0.05 being deemed significant.
Production of nano-sheets of reduced graphene oxide (rgo)
Graphene oxide (GO) was created from graphite flakes using a modified version of Hummer's process [9, 10]. In a 500 ml flask, 80 ml of sulphuric acid (H₂SO₄) was combined with 2 g of graphite flakes and 1.5 g of sodium nitrate (NaNO₃). The mixture was agitated in an ice bath for sixty minutes. Potassium permanganate (KMnO₄) (15 g) was gradually added to the suspension while being vigorously stirred, keeping the reaction temperature below 20 °C. Following the initiation of the KMnO₄, the mixture was stirred for 24 h at room temperature. The mixture became progressively pasty as the reaction grew, and the color changed from dark to light brown. After that, 150 ml of double-distilled water (DDW) and 5 ml of hydrogen peroxide (30% H₂O₂) were gradually added to the mixture while being constantly stirred, resulting in a light brown to yellow color. The combination was repeatedly and thoroughly cleaned with double-distilled water to get rid of extra KMnO₄ before being purified. Following washing, the mixture was centrifuged at 12,000 rpm for 10 min. The centrifuged GO was used to prepare the rGO. The GO was reduced using a straightforward hydrothermal method. Briefly, GO was ultrasonically sonicated into a homogenous dispersion in DDW (4 mg/ml of GO), and the resulting solution was then heated for 10 h at 160 °C in a hydrothermal autoclave. To reduce the water content and prevent rGO agglomeration, the reaction was allowed to cool to room temperature once it was finished, and the resultant product (rGO) was lyophilized for 48 h [11–13]. Various characterization techniques were used to characterize the as-synthesized materials. The Rigaku D/Max-2500 X-ray diffraction (XRD) was used to verify the phase analysis of GO and rGO.
Assortment and processing of nasopharyngeal sars-cov-2 coronavirus
A total of 10 nasopharyngeal samples were collected at Dr. D. Y. Patil Medical College, Hospital, and Research Centre Kolhapur, after getting permission from the Institutional Ethics Committee. Nasopharyngeal specimens were collected by the posterior pharynx swab, to maintain optimum viability of the virus, collected swabs were inoculated in a Hi-Viral Transport medium (Hi Media Pvt. Ltd,) and directly transported to the laboratory at 2-4 ℃. Throughout the trials, standard COVID-19 safety precautions (BSL-II) were observed [16, 17]. Standard safety precautions were followed using personal protective equipment (PPE kit). In accordance with the Biomedical Waste Management Policy (BMWM), all bio-waste was disposed of.
SARS-COV-2 RNA extraction and purification from nasopharyngeal swabs using reduced graphene oxide (RGO) carbon nano sheets
A 140 μl of nasopharyngeal VTM was reassigned to a fresh 1.5 ml sterilized centrifuge tube. A 200 μl lysis buffer (0.5 mmol EDTA, 10% SDS, 10 mmol Tris-HCl, pH 7.6) and 10 μl of Proteinase-K (20 mg/ml) were added and kept in a thermal bath for 10 min at 56 °C. Biological specimens were separated by cooling centrifugation for 5 min at 10,000 rcf, 4 °C. A 500 μl of 25% polyethylene glycol 8000 MW binding buffer was added. Then 100 μl of reduced graphene oxide carbon Nano sheet (rGO) was added, followed by gentle mixing to ensure uniform suspension. The suspension was allowed to settle at ambient temperature for five minutes [18]. The supernatant was extracted using an external magnet. Wash buffer (cold 70% ethanol) was used to wash the pellet two or three times. To elute the RNA, 60 μl of elution buffer (0.5 mmol EDTA, 10 mmol Tris-HCl, pH 8.0) was added, and the mixture was then gently stirred for five minutes at 56 °C [19]. After centrifuging at 13,000 rcf for 5 min, the 60 μl supernatant was collected in a fresh microcentrifuge tube and kept at-20 °C.
Extraction and purification of SARS-COV-2 RNA from oropharyngeal/nasopharyngeal swabs using commercial magnetic bead extraction method
RNA was isolated from the nasopharyngeal sample using the commercially available magnetic beads RNA extraction kit MagMAX Total Nucleic Acid Isolation Kit, thermo-fisher labs. Nasopharyngeal tissues were used to extract RNA in accordance with the manufacturer's instructions [20–22]. Each sample generated 60 μl of RNA, which was then kept at-20 °C.
RNA Purity evaluation VIA UV-VIS spectroscopy
The purity and yield of the extracted RNA were evaluated by both commercially available kits and by using rGO-assisted RNA extraction using UV-Vis spectroscopy. The A260/A280 ratio provides information about protein contamination as shown in Supplementary Table 1. The formula 1 OD260 unit = 50 μg/ml can be used to calculate the concentration of RNA [18].
Real-time PCR-SARS-COV-2
RT-PCR was carried out using both commercially and rGO-assisted isolated SARS-CoV-2 coronavirus RNA samples. The real-time PCR examination was implemented for qualitative assessment of extracted RNA. Exact primer-probe (Rnase P, N gene, and ORF-1ab gene) was added to the PCR mixture as per the manufacturer's instructions (CoviPath, Thermo Fisher). A total of 40 RT-PCR thermal cycles were performed, which included cDNA synthesis at 50 °C for 15 min, initial denaturation at 95 °C for 3 min, and annealing at 58 °C for 1 minute [23]. The entire volume of the reaction system was 25 μl.
Electrophoresis with agarose gel
The amplified PCR products from the commercial and rGO-assisted techniques were separated on a 1.5% agarose gel for further confirmation. For staining, 0.5 µg/ml of ethidium bromide was utilized. A 100 bp DNA ladder (Hi-Media, Mumbai, India) was employed. A gel imager (Applied Biosystem) was used to take pictures of the electrophoresed gel [24].
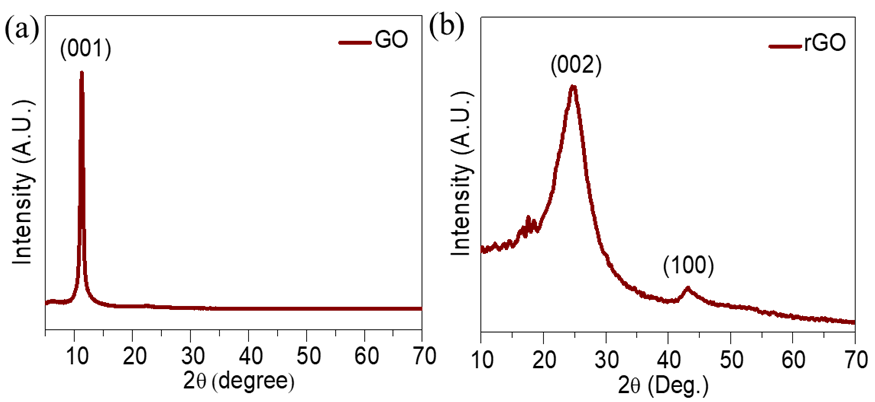
Fig. 1: XRD patterns of (a) GO and (b) rGO
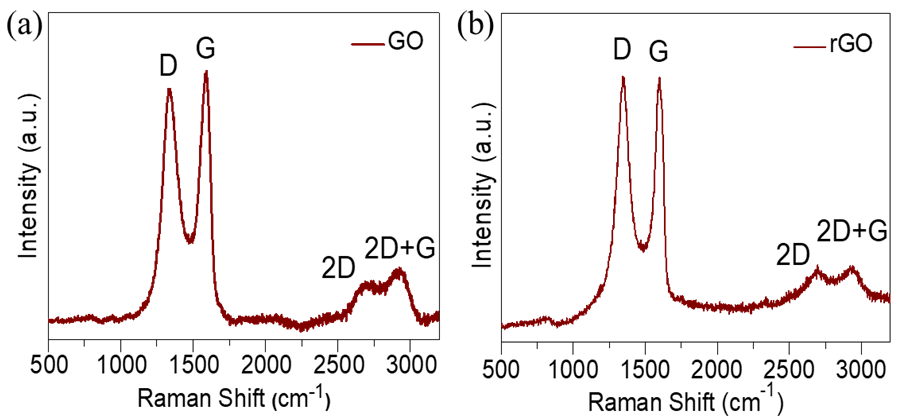
Fig. 2: Raman spectra of (a) GO and (b) rGO
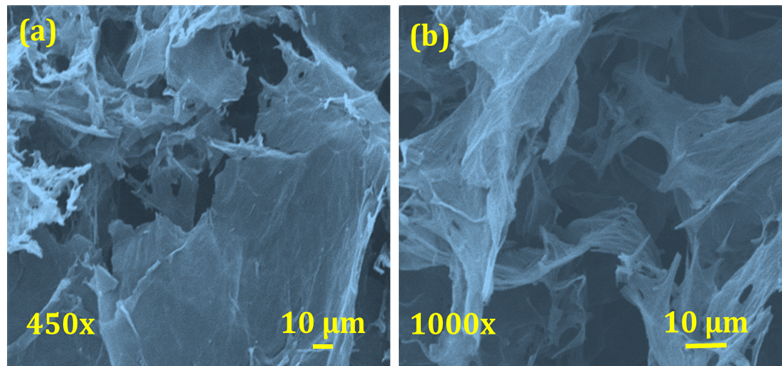
Fig. 3: (a, b) FE-SEM micrographs of rGO at different magnifications (450x and 1000x)
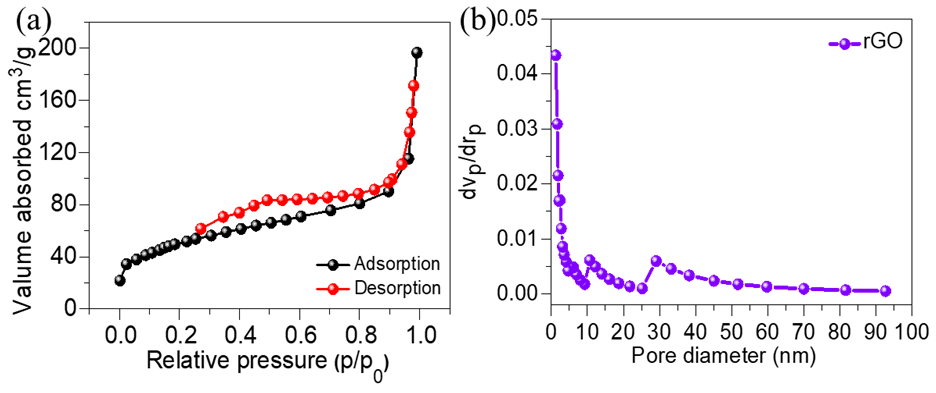
Fig. 4: (a) N2 adsorption/desorption isotherm, and (b) BJH pore size distribution plot of rGO
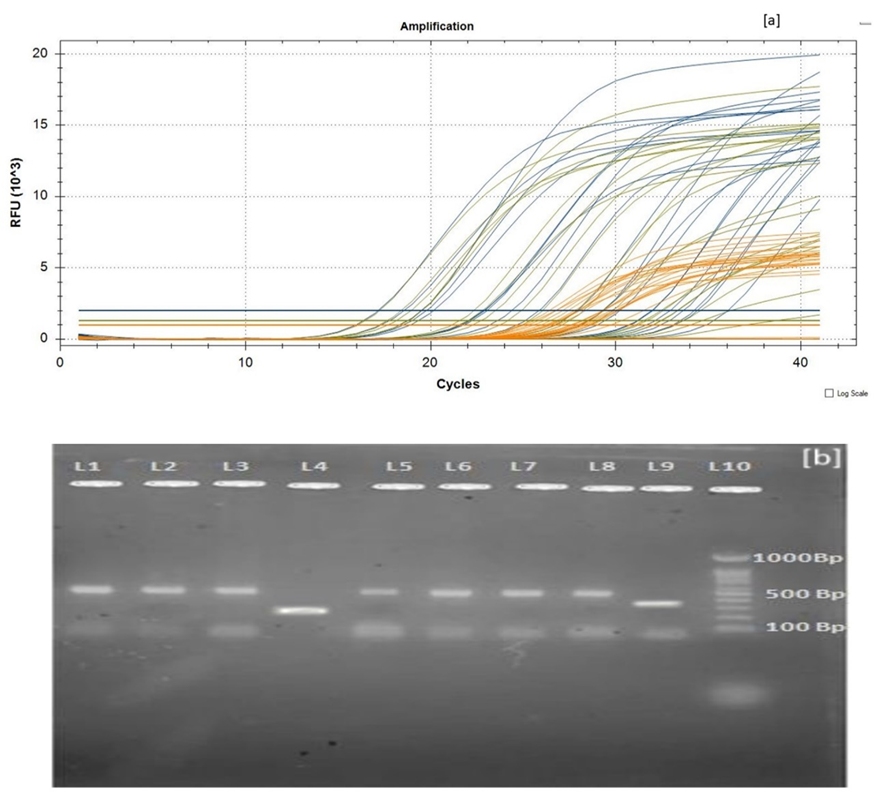
Fig. 5: A. rGO carbon Nano sheet mediated SARS-CoV-2 RNA-RT PCR gene amplification curve B: Agarose gel electrophoresis of obtained RT-PCR product

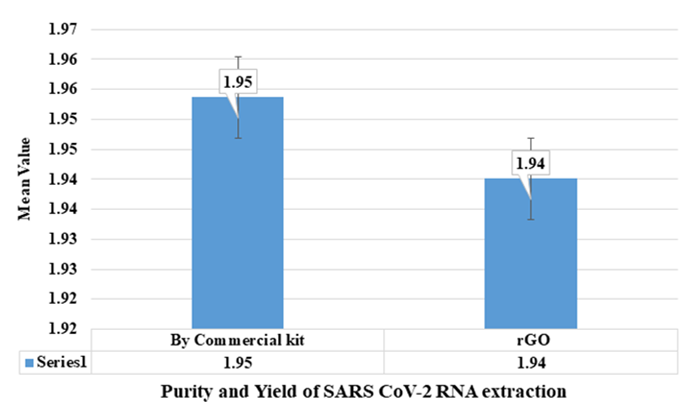
Fig. 6: Purity and Yield of SARS-CoV-2 RNA extraction with commercial kit
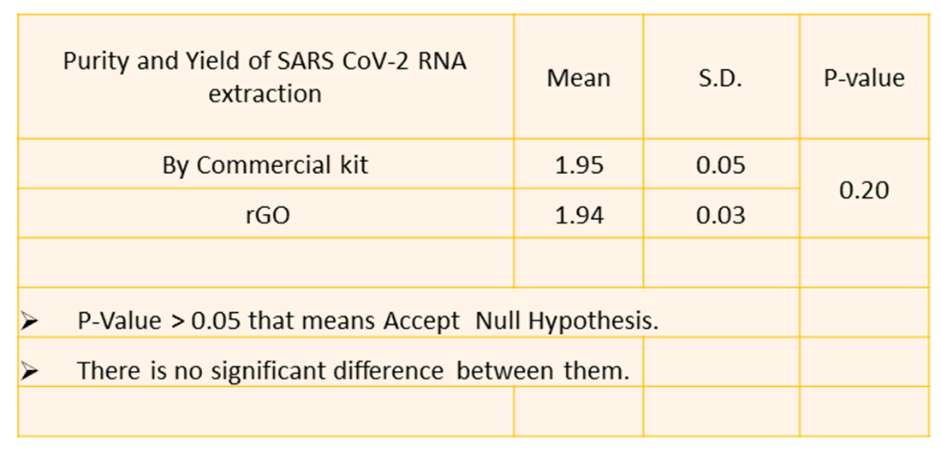
Fig. 7: Statistical significance of p-value
RESULTS
XRD study
Fig. 1(a, b) displays the X-ray diffraction (XRD) patterns of GO and rGO. The complete oxidation of graphite particles is shown by the large characteristic peak in the XRD of GO at 11.1°, which corresponds to the (001) plane. After reduction, the XRD pattern of rGO displays diffraction peaks at 24.51° and 42.89°, which correspond to the (002) and (100) planes, respectively. The appearance of the scattered peak at approximately 24.51° further confirms the creation of multi-layered rGO nanostructures and the decrease of GO.
Raman analysis
Raman spectroscopy is instrumental in analyzing defects and characterizing graphene layers. The Raman spectrum of rGO, shown in fig. 2 (c, d), exhibits three major peaks: The D, G, and 2D bands are located at 1345, 1591, and 2697 cm⁻¹, respectively. The vibrations of sp³ disordered carbon atoms are linked to the D band, while the G band is indicative of the sp² hybridized carbon atoms in graphite. Additionally, a D+G band at 2935 cm⁻¹ is observed, corresponding to disorder-induced scattering. The 2D band’s presence confirms the multi-layered architecture of both GO and rGO. The D/G intensity ratio (I_D/I_G) of GO and rGO were 0.99 and 1.01, respectively. The slight increase in the D/G ratio after reduction suggests the incorporation of defects during the hydrothermal reduction process. This analysis confirms that the hydrothermal approach successfully reduced GO to rGO, with an increased disorder in the graphene structure.
FE-SEM study
Using field emission scanning electron microscopy (FE-SEM), the morphology of the rGO electrode was examined. As seen in fig. 3 (a, b), the rGO Nano sheets displayed a distinct 3D porous structure with interconnected sheets at low (450x) and high (1000x) magnifications. As seen in fig. 2(c), the network's pore sizes varied from several micrometers to sub-micrometers. Thin layers of stacked graphene sheets made up the walls of the pores, and physical cross-linking inside the rGO framework was caused by partial overlapping or collapse of the flexible graphene sheets.
Bet analysis
The nitrogen (N₂) adsorption-desorption isotherm of rGO is displayed in fig. 4(a), and the matching BJH pore-size distribution map is shown in fig. 4(b). The H3-type hysteresis loop observed in the isotherm suggests that rGO has a mesoporous structure, which is characteristic of materials that facilitate both absorption and desorption processes. The rGO material has an average pore size of 7.15 nm and a significant surface area of 225 m²/g. The mesoporous structure of rGO contributes to enhanced ion diffusion and low resistance paths, making it well-suited for high-performance, ultrasensitive detection applications.
Validation of extracted RNA
The RNA extracted using rGO carbon Nano sheets was quantified by UV-Vis spectroscopy, with the results showing that the rGO-based extraction yielded significantly higher RNA concentrations compared to the commercial method. Table 1 compares the RNA yield and purity between the rGO Nano sheet-based method and traditional kit-based RNA extraction. The rGO extraction method showed a marked increase in RNA concentration, with a higher purity profile, further validating the efficiency of rGO as an RNA extraction material.
Statistical analysis of RNA yield
Statistical analysis of RNA yields (table 1) showed no significant difference between the rGO and commercial extraction methods (p-value>0.05), indicating that rGO Nano sheets are a superior material for extracting RNA from clinical samples. The increased yield with rGO was consistent across multiple sample types, demonstrating the robustness of the rGO-based extraction protocol as shown in fig. 6 and tables 1-3. There is a significant increase in RNA yield (1.959 ng/µl) and improved sensitivity in RT-PCR, further validating the advantage of using rGO for RNA extraction shown in fig. 7.
Table 1: RNA extraction and quantification of results by bio-spectrophotometer
| Number of experiments | Purity and yield of SARS CoV-2 RNA (ng/µl) extraction by commercial kit |  Purity and Yield of SARS CoV-2 RNA extraction by rGO Purity and Yield of SARS CoV-2 RNA extraction by rGO |
| 1 | 1.877 | 1.871 |
| 2 | 1.881 | 1.879 |
| 3 | 1.866 | 1.919 |
| 4 | 1.976 | 1.971 |
| 5 | 1.980 | 1.918 |
| 6 | 1.972 | 1.962 |
| 7 | 1.981 | 1.959 |
| 8 | 1.976 | 1.961 |
| 9 | 1.984 | 1.962 |
| 10 | 1.979 | 1.970 |
| 11 | 1.983 | 1.928 |
| 12 | 1.970 | 1.962 |
| 13 | 1.972 | 1.959 |
Table 2: Comparison of Cq value of RT-PCR between commercial RNA extraction kit and reduced graphene oxide nano sheet (NA-No amplification, PC-Positive control, NC-Negative control, rGO-reduced graphene oxide, com. kit-commercial RNA extraction kit)
| Sample ID | Kit | ORF lab | N gene | R-nase P |
| S1 | rGO/com. Kit | 20.17/36.06 | 28.29/25.96 | 17.66/32.44 |
| S2 | rGO/com. Kit | 18.57/32.01 | 27.93/26.22 | 34.70/13.68 |
| S3 | rGO/com. Kit | 33.65/32.95 | 28.48/26.30 | 16.48/21.57 |
| S4 | rGO/com. Kit | 17.18/23.06 | 27.54/27.64 | 24.17/27.16 |
| S5 | rGO/com. Kit | 24.39/27.95 | 26.85/26.44 | 22.74/28.52 |
| S6 | rGO/com. Kit | 22.93/29.31 | 29.70/26.44 | 27.93/26.48 |
| S7 | rGO/com. Kit | 34.40/29.30 | 27.93/26.47 | 27.93/26.48 |
| S8 | rGO/com. Kit | 25. 36/19.50 | 28.67/24.88 | 25.33/18.16 |
| S9 | rGO/com. Kit | 31.96/30.22 | 25.55/25.28 | 31.52/29.09 |
| S10 | rGO/com. Kit | 22.88/19.78 | 28.13/25.05 | 22.04/18.81 |
| NC | rGO/com. Kit | NA | NA | NA |
| PC | rGO/com. Kit | 34.23 | 33.67 | NA |
Table 3: Comparison of extraction of SARS-CoV-2 RNA using nanoparticles and their limitations and techniques used
| S. No. | Sample | Nano material use | Limitation | Sensitivity | Detection method | Technique use | Ref. |
| 1 | Nasal swab and saliva samples | Carbon nanotube-based extraction | (LoQ) of 6.4 copies/μl in PBS buffer and LoQ of 9.2 copies/ μl in 50% human saliva |
PCR sensitivity LoQ of 6.4 copies/μl in PBS | RT-qPCR | Capture ssDNA sequences attached to carbon nanotubes |
[41] |
| 2 | Nasal swab and saliva samples | ACE2-SWCNTs based Nano sensor | ∼104−106 viral copies per μl |
∼101−104 viral copies per μl translating to ∼0.005−5 pM S RBD | Biosensor | ACE2-single walled carbon nanotubes | [42] |
| 3 | Nasal swab and saliva samples | carbon electrode composed of AuNP array | 1.0 pg ml-1 | 0.001 to 100 ng ml-1 | Electrochemical chip | Carbon nanotube with gold array | [43] |
| 5 | SARS CoV-2 | Magnetic Nanoparticles functionalized with carboxyl group | Protocol useful for SARS-CoV-2RNA extraction however, confirmatory results of different experimental study are lacking | Not specified | Not specified | Magnetic Bead with RT qPCR and Lamp | [44] |
| 6 | SARS CoV-2 RNA | rGO Nano sheets | A protocol by using rGO nanosheets need to be validated for commercial kit purpose | 100% sensitivity was obtained for gene, N gene and RDRP gene specific SARS-CoV-2 virus | RT-qPCR | rGO Nano sheets | Present study |
Agarose gel electrophoresis with RT-PCR
Real-time PCR assay was implemented on RNA samples extracted using rGO carbon Nano sheets, targeting the R-nase P, N gene, and ORF-1ab genes of SARS-CoV-2. The RT-PCR results, shown in fig. 5(A), demonstrated a quick and strong amplification of the targeted genes, confirming the occurrence of the SARS-CoV-2 genome. The amplification peaks were observed after just 15 cycles of RT-PCR, indicating efficient RNA quality and successful amplification. These results were further validated by agarose gel electrophoresis (fig. 5(B), where distinct cDNA bands appeared corresponding to the amplified SARS-CoV-2 genes. Ethidium bromide staining revealed significant fluorescence, confirming the successful extraction and amplification of SARS-CoV-2 cDNA. In table 2, the CQ values (Cycle Quantification values) are reported for the different genetic markers (ORF lab, N gene, and R-nase P) in each sample. The number of PCR cycles needed for the fluorescent signal to surpass the threshold and signify the CQ value represents the detection of the target nucleic acid. Two values per marker are listed, which likely translate to two distinct replicates or outcomes for the same sample. For example, for Sample S1 under the ORF lab column, you have: 20.17/36.06 (CQ values for the two replicates). Sample ID: The identifier for each sample (e. g., S1, S2, etc.). Kit: The specific kit used for testing (e. g., rGO/com. Kit). ORF lab: CQ values for the ORF lab gene, with two values per sample representing two separate assays or replicates’ gene: CQ values for the N gene, similarly with two values per sample. R-nase P: CQ values for the R-nase P gene, again with two values per sample for the Negative Control (NC) and Positive Control (PC): NC: NA (No amplification expected; typically, should not show any CQ value). PC: For the positive control, only two CQ values are reported for ORF and N gene. R-nase P does not have a value listed, which may suggest that the R-nase P marker was not assessed or was unnecessary for the positive control. The CQ values that are reported as "NA" (Not Applicable) could indicate that amplification was not detected, or the test was not performed for that marker. Lower CQ values indicate higher initial template concentration. Higher CQ values suggest less template and possibly lower detection sensitivity.
Advantages of RGO nano sheets
The rGO Nano sheets detects higher Sensitivity approach in Sample 6 offers 100% sensitivity for detecting the SARS-CoV-2 virus, outperforming other techniques in terms of sensitivity, especially for the N gene and RDRP gene. This makes rGO a promising material for viral RNA detection at low concentrations. Compatibility with RT-PCR. The rGO-based method uses RT-PCR, which is a well-established and reliable technique for viral detection, adding a layer of confidence in the results. Broad Detection of Multiple genes with rGO Nano sheets are effective in detecting multiple viral genes, ensuring robust results across various targets, such as the N gene and RDRP gene, which are critical for identifying SARS-CoV-2. Despite the need for validation, the rGO method holds promise for future integration into commercial kits, potentially offering a more versatile and accurate detection system.
LIMITATIONS OF RGO
Although the sensitivity is promising, further validation and optimization of protocols for different sample types (e.g., saliva, nasal swabs, dry swab) and under real-world conditions are essential to confirm its robustness and commercial viability.
DISCUSSION
The rGO was synthesized and characterized by XRD, Raman, FE-SEM, and BET analysis. The rGO Nano sheets have a well-defined 3D porous structure and interconnected Nano sheets. The network's pore walls featured thin layers of ordered graphene sheets, and the network's pore size varied from several micrometers to sub-micrometers. The physical cross-linking points in the rGO framework caused flexible graphene sheets to partially overlap or collapse. A mesoporous structure of rGO, a characteristic of porous materials, allows for absorption and desorption. Furthermore, the mesoporous structure of rGO can help in the reduction of ion diffusion channels and low resistance paths, thereby matching the requirements of high-value ultrasensitive detection applications. The significant RNA extraction efficacy of rGO carbon Nano sheets was observed compared to the commercial RNA extraction kit. The purity of viral RNA extracted by the commercial kit is equivalent to rGO-based carbon Nano sheets (table 1). Extracted RNA (rGO carbon Nano sheet and commercial RNA extraction kit) was amplified by performing RT-PCR. Comparing the rGO carbon Nano-sheet mediated extraction to the commercial RNA extraction kit, an equivalent RNA yield was found. Results obtained from our study compared with previous studies (table 3) are comparable. Consequently, rGO Nano sheet-based RNA extraction may be a substitute technique for removing SARS-CoV-2 RNA from biological samples. The rGO Nano sheet ribonucleic acid extraction uses the lysis and binding procedures, and extracted pure RNA can be further used for PCR-based amplification and identification. Magnetic adsorption techniques using rGO Nano sheet viral RNA extraction were accomplished by a rapid and simple low-cost RNA extraction method from nasopharyngeal swabs [25–27]. GO has shown higher stability in water [28–30]. There is an interaction between polar groups (non-covalent) and the formation of hydrogen bonds between GO and single-stranded nucleic acids such as ribonucleic acid (RNA) [16, 31, 32]. rGO Nano sheets were utilized for both solid-phase support and magnetic separation in order to adsorb RNA [33-35]. The rGO responds better to the applied magnetic field and has a larger surface area for RNA molecule adsorption [36-39]. The positively charged rGO Nano sheet surface adsorbs negatively charged RNA molecules [40–42]. Phosphodiester bonds with hydroxyl and amino groups form the backbone of RNA [46, 47]. It forms hydrogen bonds with molecules that include hydroxyl, carboxyl, phosphate, sulfate, Mg++, and Ca++groups [48]. RNA is given a negative charge by the phosphate groups present in phosphodiester linkages. Further to ensure robustness, further validation is needed in diverse real-world settings. This includes testing across a wide range of sample types (e. g., nasal swabs, saliva, blood) and assessing the method's performance with different viral strains [49]. Comparing the method's performance with other RNA extraction techniques in these diverse contexts will be crucial. Additionally, evaluating the method’s cost-effectiveness and ease of use for point-of-care testing will further support its widespread adoption. By addressing these improvements, the rGO-based RNA extraction method could be developed into a versatile and commercially viable tool for pathogen detection, providing a significant advance in diagnostic technology.
CONCLUSION
In this study, we successfully extracted pure RNA using rGO Nano sheets, with quantitative confirmation of RNA presence. The RNA adsorption onto rGO Nano sheets is facilitated by hydrogen bonding and electrostatic interactions. Extracted RNA was successfully amplified through RT-PCR and confirmed via agarose gel electrophoresis. The results from this method were comparable to those obtained with a commercially available RNA extraction kit. There is a significant increase in RNA yield (1.959 ng/µl) and improved sensitivity in RT-PCR, further validating the advantage of using rGO for RNA extraction. These findings suggest that the rGO carbon Nano nanosheet-based RNA extraction method holds substantial potential for the detection of COVID-19, offering an efficient alternative to traditional methods. Beyond COVID-19, the rGO Nano sheet method has significant potential for multiplexed pathogen detection, which would allow the detection of several diseases (such as bacteria or viruses) at once from a single sample, enhancing the diagnostic capability of the method.
ACKNOWLEDGEMENT
The authors appreciate the support from the Intramural project: DYPES/DU/R&D2021/279, D. Y. Patil Education Society (Deemed to be University), Kolhapur-416006, Maharashtra, India.
FUNDING
Nil
AUTHORS CONTRIBUTIONS
CONFLICT OF INTERESTS
The authors have declared that they have no conflict of interest.
REFERENCES
Kashte S, Gulbake A, El Amin SF, Gupta A. COVID-19 vaccines: rapid development implications challenges and future prospects. Hum Cell. 2021;34(3):711-33. doi: 10.1007/s13577-021-00512-4, PMID 33677814.
Gupta A, Shivaji K, Kadam S, Gupta M, Rodriguez HC, Potty AG. Immunomodulatory extracellular vesicles: an alternative to cell therapy for COVID-19. Expert Opin Biol Ther. 2021;21(12):1551-60. doi: 10.1080/14712598.2021.1921141, PMID 33886388.
WHO. Coronavirus (COVID-19) Dashboard. World Health Organization. Available from: https://covid19.who.int. [Last accessed on 27 Sep 2022].
Klein S, Muller TG, Khalid D, Sonntag Buck V, Heuser AM, Glass B. SARS-CoV-2 RNA extraction using magnetic beads for rapid large scale testing by RT-qPCR and RT-LAMP. Viruses. 2020 Aug 1;12(8):863. doi: 10.3390/v12080863, PMID 32784757.
Zhao Z, Cui H, Song W, Ru X, Zhou W, Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. doi: 10.1101/2020.02.22.961268.
Parihar A, Puranik N, Parihar DS, Ranjan P, Khan R. Currently available biosensor-based approaches for severe acute respiratory syndrome corona virus 2 detection. Adv Biosens Virus Detect Smart Diagn Combat SARS-CoV. 2022 Jan 1;2:373-90. doi: 10.1016/B978-0-12-824494-4.00016-3.
Singh V, Batoo KM, Singh M. Fabrication of chitosan-coated mixed spinel ferrite integrated with graphene oxide (GO) for magnetic extraction of viral RNA for potential detection of SARS-CoV-2. Appl Phys A Mater Sci Process. 2021 Dec 1;127(12):960. doi: 10.1007/s00339-021-05067-7, PMID 34866806.
Somvanshi BS, B Kharat P, S Saraf T, B Somwanshi S, B Shejul S, M Jadhav K. Multifunctional nano-magnetic particles assisted viral RNA-extraction protocol for potential detection of COVID-19. Mater Res Innov. 2020;25(3):1-6. doi: 10.1080/14328917.2020.1769350.
Zambry NS, Obande GA, Khalid MF, Bustami Y, Hamzah HH, Awang MS. Utilizing electrochemical based sensing approaches for the detection of sars-cov-2 in clinical samples: a review. Biosensors. 2022;12(7):473. doi: 10.3390/bios12070473, PMID 35884276.
Mohamed MJ, Bhat DK. Novel ZnWO4/RGO nanocomposite as high performance photocatalyst. AIMS Mater Sci. 2017;4(1):158-71. doi: 10.3934/matersci.2017.1.158.
Ahmed B, Ojha AK, Hirsch F, Fischer I, Patrice D, Materny A. Tailoring of enhanced interfacial polarization in WO3 nanorods grown over reduced graphene oxide synthesized by a one step hydrothermal method. RSC Adv. 2017;7(23):13985-96. doi: 10.1039/C7RA00730B.
Bharath G, Anwer S, Mangalaraja RV, Alhseinat E, Banat F, Ponpandian N. Sunlight-induced photochemical synthesis of Au nanodots on α-Fe2O3@Reduced graphene oxide nanocomposite and their enhanced heterogeneous catalytic properties. Sci Rep. 2018 Dec 1;8(1):5718. doi: 10.1038/s41598-018-24066-y.
Iskandar F, Abdillah OB, Stavila E, Aimon AH. The influence of copper addition on the electrical conductivity and charge transfer resistance of reduced graphene oxide (rGO). New J Chem. 2018;42(19):16362-71. doi: 10.1039/x0xx00000x.
Dresselhaus MS, Jorio A, Hofmann M, Dresselhaus G, Saito R. Perspectives on carbon nanotubes and graphene raman spectroscopy. Nano Lett. 2010;10(3):751-8. doi: 10.1021/nl904286r, PMID 20085345.
Xu Y, Sheng K, Li C, Shi G. Self-assembled graphene hydrogel via a one step hydrothermal process. ACS Nano. 2010 Jul 27;4(7):4324-30. doi: 10.1021/nn101187z, PMID 20590149.
Parra Guardado AL, Sweeney CL, Hayes EK, Trueman BF, Huang Y, Jamieson RC. Development of a rapid pre-concentration protocol and a magnetic bead-based RNA extraction method for SARS-CoV-2 detection in raw municipal wastewater. Environ Sci (Camb). 2022 Jan 1;8(1):47-61. doi: 10.1039/D1EW00539A.
Kerachian MA, Amel Jamehdar SA, Azghandi M, Keyvanlou N, Mozaffari Jovin S, Javadmanesh A. Developing novel liquid biopsy by selective capture of viral RNA on magnetic beads to detect COVID-19. Iran J Basic Med Sci. 2022 Jun 1;25(6):762-6. doi: 10.22038/IJBMS.2022.65260.14379, PMID 35949306.
Hernandez S, Cardozo F, Myers DR, Rojas A, Waggoner JJ. Simple and economical RNA extraction and storage packets for viral detection from serum 1 or plasma; 2022. doi: 10.1101/2022.01.28.22270041.
Ngamsom B, Iles A, Kamita M, Kimani R, Rodriguez Mateos P, Mungai M. An integrated lab-on-a-chip device for RNA extraction amplification and CRISPR-Cas12a-assisted detection for COVID-19 screening in resource-limited settings. medRxiv. 2022. doi: 10.1101/2022.01.06.22268835.
Possebon FS, Ullmann LS, Malossi CD, Miodutzki GT, Da Silva EC, Machado EF. A fast and cheap in-house magnetic bead RNA extraction method for COVID-19 diagnosis. J Virol Methods. 2022 Feb 1;300:114414. doi: 10.1016/j.jviromet.2021.114414, PMID 34896456.
Phan T, Stephenson R, Cai T, Andacic N, McKew G. A comparison of SARS-CoV-2 RNA extraction with the quickgene-810 nucleic acid isolation system compared to the EZ1 advanced DSP virus kit. Access Microbiol. 2022 May 27;4(5):acmi000353. doi: 10.1099/acmi.0.000353, PMID 36003356.
Shen J, Hartmann EM. DNA extraction protocol for low biomass environmental samples: adapted from the lucigen masterpure complete DNA and RNA Purification Kit manual; 2021. doi: 10.21203/rs.3.pex-1658/v1.
Chaudhary S, Patel D, Vyas K, Chaudhary P, Singhania P, Patel J. SARS-CoV-2 direct real time PCR without RNA extraction: appropriate method or not? European Journal of Medical and Health Sciences. 2022 Jan 27;4(1):41-7. doi: 10.24018/ejmed.2022.4.1.1164.
Ali TH, Mandal AM, Heidelberg T, Hussen RS. Sugar-based cationic magnetic core shell silica nanoparticles for nucleic acid extraction. RSC Adv. 2022 May 5;12(22):13566-79. doi: 10.1039/d2ra01139e, PMID 35530382.
Poon LL, Chan KH, Wong OK, Yam WC, Yuen KY, Guan Y. Early diagnosis of SARS coronavirus infection by real-time RT-PCR. J Clin Virol. 2003;28(3):233-8. doi: 10.1016/j.jcv.2003.08.004, PMID 14522060.
Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020. doi: 10.1101/2020.02.26.20028373.
Wozniak A, Cerda A, Ibarra Henriquez C, Sebastian V, Armijo G, Lamig L. A simple RNA preparation method for SARS-CoV-2 detection by RT-qPCR. Sci Rep. 2020 Dec 1;10(1):16608. doi: 10.1038/s41598-020-73616-w, PMID 33024174.
Ambrosi C, Prezioso C, Checconi P, Scribano D, Sarshar M, Capannari M. SARS-CoV-2: comparative analysis of different RNA extraction methods. J Virol Methods. 2021 Jan 1;287:114008. doi: 10.1016/j.jviromet.2020.114008, PMID 33160015.
Won J, Lee S, Park M, Kim TY, Park MG, Choi BY. Development of a laboratory-safe and low cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19). Exp Neurobiol. 2020;29(2):107-19. doi: 10.5607/en20009, PMID 32156101.
Grant PR, Turner MA, Yen Shin G, Nastouli E, Levett LJ. Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. BioRxiv. 2020 Apr 9. doi: 10.1101/2020.04.06.028316.
Whitney ON, Kennedy LC, Fan VB, Hinkle A, Kantor R, Greenwald H. Sewage salt silica and SARS-CoV-2 (4S): an economical kit free method for direct capture of SARS-CoV-2 RNA from wastewater. Environ Sci Technol. 2021 Apr 20;55(8):4880-8. doi: 10.1021/acs.est.0c08129, PMID 33759506.
Wee SK, Sivalingam SP, Yap EP. Rapid direct nucleic acid amplification test without RNA extraction for SARS-COV-2 using a portable PCR thermocycler. Genes (Basel). 2020 Jun 1;11(6):664. doi: 10.3390/genes11060664, PMID 32570810.
Barza R, Patel P, Sabatini L, Singh K. Use of a simplified sample processing step without RNA extraction for direct SARS-CoV-2 RT-PCR detection. J Clin Virol. 2020 Nov 1;132:104587. doi: 10.1016/j.jcv.2020.104587, PMID 32898817.
Perez Cataluna A, Cuevas Ferrando E, Randazzo W, FalcoI, Allende A, Sanchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci Total Environ. 2021 Mar 1;758:143870. doi: 10.1016/j.scitotenv.2020.143870, PMID 33338788.
Mahmoud SA, Ganesan S, Ibrahim E, Thakre B, Teddy JG, Raheja P. Evaluation of RNA extraction-free method for detection of SARS-CoV-2 in salivary samples for mass screening for COVID-19. BioMed Res Int. 2021;2021:5568350. doi: 10.1155/2021/5568350, PMID 34327228.
Rahimpour E, Lotfipour F, Jouyban A. A minireview on nanoparticle-based sensors for the detection of coronaviruses. Bioanalysis. 2021 Dec 1;13(24):1837-50. doi: 10.4155/bio-2021-0006, PMID 34463130.
Tang C, He Z, Liu H, Xu Y, Huang H, Yang G. Application of magnetic nanoparticles in nucleic acid detection. J Nanobiotechnology. 2020;18(1):62. doi: 10.1186/s12951-020-00613-6, PMID 32316985.
Rodriguez Diaz C, Lafuente Gomez N, Coutinho C, Pardo D, Alarcon Iniesta H, Lopez Valls M. Development of colorimetric sensors based on gold nanoparticles for SARS-CoV-2 RdRp, E and S genes detection. Talanta. 2022 Jun 1;243:123393. doi: 10.1016/j.talanta.2022.123393, PMID 35325745.
Owida HA, Al Nabulsi JI, Turab NM, Louzi N. Nanotechnology role development for COVID-19 pandemic management. J Nanotechnol. 2022;1:2-12. doi: 10.1155/2022/1872933open_in_new.
Dighe K, Moitra P, Alafeef M, Gunaseelan N, Pan D. A rapid RNA extraction free lateral flow assay for molecular point of care detection of SARS-CoV-2 augmented by chemical probes. Biosens Bioelectron. 2022 Mar 15;200:113900. doi: 10.1016/j.bios.2021.113900, PMID 34959185.
Jeong S, Gonzaez Grandio E, Navarro N, Pinals RL, Ledesma F, Yang D. Extraction of viral nucleic acids with carbon nanotubes increases SARS-CoV-2 quantitative reverse transcription polymerase chain reaction detection sensitivity. ACS Nano. 2021;15(6):10309-17. doi: 10.1021/acsnano.1c02494, PMID 34105936.
Pinals RL, Ledesma F, Yang D, Navarro N, Jeong S, Pak JE. Rapid SARS-CoV-2 spike protein detection by carbon nanotube based near infrared nanosensors. Nano Lett. 2021 Mar 10;21(5):2272-80. doi: 10.1021/acs.nanolett.1c00118, PMID 33635655.
Layqah LA, Eissa S. An electrochemical immunosensor for the corona virus associated with the middle-east respiratory syndrome using an array of gold nanoparticle modified carbon electrodes. Mikrochim Acta. 2019 Apr 1;186(4):224. doi: 10.1007/s00604-019-3345-5, PMID 30847572.
Klein S, Muller TG, Khalid D, Sonntag Buck V, Heuser AM, Glass B. SARS-CoV-2 RNA extraction using magnetic beads for rapid large scale testing by RT-qPCR and RT-LAMP. Viruses. 2020;12(8):863. doi: 10.3390/v12080863, PMID 32784757.
Kaboli MA, Hashim AA, Dhiya Altememy, Javad Saffari Chaleshtori, Mehdi Rezaee, Sayedeh Azimeh Hosseini, Pegah Khosravian. Silk fibroin-coated mesoporous silica nanoparticles enhance 6-thioguanine delivery and cytotoxicity in breast cancer cells. Int J Appl Pharm. 2025;17(1):275-83. doi: 10.22159/ijap.2025v17i1.52882.
Sharma T, Sharma A. An emerging era in drug delivery system for treatment of malaria: wave from conventional to advanced technology. Int J Appl Pharm. 2025;17(1):48-58. doi: 10.22159/ijap.2025v17i1.52285.
Eguilaz M, Villalonga R, Rivas G. Electrochemical biointerfaces based on carbon nanotubes mesoporous silica hybrid material: bioelectrocatalysis of hemoglobin and biosensing applications. Biosensors and Bioelectronic. 2018 Jul 15;111:144-51. doi: 10.1016/j.bios.2018.04.004.
Barik G, Sarkar I, Biswas D, Sahoo SK. Dueal target reverse transcription polymerase chain reaction for SARS-COV-2 for enhancing accuracy with E gene and ORF 1B gene: a cross sectional study. Asian J Pharm Clin Res. 2024;17(12):211-21. doi: 10.22159/ajpcr.2024v17i12.53318.
Suvarna NH, Mathew JE, Raj V, Harees S, kumar L, Verma R. Integrative Qsar analysis of oxadiazole derivatives resolving molecular determinants for anti-tubercular activity and rational drug design. Int J Pharm. 2024;16(5):157-65. doi: 10.22159/ijap.2024v16i5.51468.