
Int J Pharm Pharm Sci, Vol 17, Issue 9, 52-62Original Article
COMPARATIVE MOLECULAR DOCKING STUDIES OF 7-HYDROXY COUMARIN DERIVATIVES
TYAGI ALKAa*, SADGIR PRIYANKAb
a*Department of Pharmaceutical Chemistry, Banasthali Vidyapith, Rajasthan, India. bDepartment of QA, Shri. Pandit Baburao Chughule College of Pharmacy, Thane, India
*Corresponding author: Tyagi Alka; *Email: alka.tyagi35@gmail.com
Received: 03 Apr 2025, Revised and Accepted: 11 Jun 2025
ABSTRACT
Objective: The derivatives of 7-hydroxycoumarin were studied as novel human acetylcholinesterase (AChE) inhibitors that could be useful in the treatment of Alzheimer's disease (AD).
Methods: This study set out to determine the lead molecules (synthesized 7-hydroxycoumarin derivatives) and investigate the viability of two distinct docking strategies for our target AChE: Auto Dock vina and Schrodinger Glide. Using these tools, we have conducted a comparative analysis of ligand binding affinity and binding poses.
Results: Among all the Coumarin-derived compounds docked against hAChE, 2 compounds, namely, 2-((2-oxo-2H-chromen-4-yl) oxy)-N-(pyridin-3-yl) acetamide derivative 4(d) and 7-(2-oxo-2-(10 H-phenothiazin-10-yl) ethoxy)-2H-chromen-2-one, 4(f) showed the best docking results.
Conclusion: Schrodinger was found to be comparatively more helpful in blind docking pose prediction and consistently outperformed another program. In the current study, the most powerful compound’s interactions with amino acids in binding pockets are analysed and compared using both docking tools.
Keywords: Alzheimer’s disease, Docking, In silico, Coumarin, AChE, 7-hydroxy coumarin
© 2025 The Authors. Published by Innovare Academic Sciences Pvt Ltd. This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/)
DOI: https://dx.doi.org/10.22159/ijpps.2025v17i9.54600 Journal homepage: https://innovareacademics.in/journals/index.php/ijpps
INTRODUCTION
Humans' typical character is determined by cognition, which is a blend of consciousness, perception, familiarity, skills, memories, introspection, planning, and judgment [1–5]. Numerous factors, including aging, chemical exposure, brain trauma, and the onset of neurodegenerative diseases like schizophrenia, depression [6], and Alzheimer's disease (AD), are linked to memory loss and cognitive impairment [7–8]. Prominent characteristics of AD include the buildup of defective mitochondria and neuronal autophagy impairment, which results in impaired mitophagy [9]. For millions of elderly people, dementia is the most common cause of memory loss and cognitive decline [10-11]. The neurotransmitters that regulate cognitive function are acetylcholine (ACh), dopamine, serotonin, and glutamate [12, 13]. The loss of cholinergic system presynaptic indicators in the cortical area of the brain, which is crucial for memory and learning processes, is a hallmark of AD. The genesis of AD is assumed to be influenced by a number of variables, including low Ach levels, accumulation of abnormal proteins in brains, oxidative stress, tau protein hyperphosphorylation, and biometal imbalance [14]. AD is characterized by neuronal cell death due to the amplification of aberrant proteins, such as β-amyloid plaques (Aβ) and neurofibrillary tangles [15] in the affected individuals' brain [16]. There are three licensed AChE inhibitors currently used to fight Alzheimer's disease, viz., Donepezil, Galantamine, and Rivastigmine (fig 1), as well as NMDA antagonist, Memantine [16-18]. The enzyme cavity of AChE is a narrow, 20Å deep groove that contains two binding sitescatalytic active site (CAS) and the Peripheral anionic site (PAS) [19]. Molecular docking, the strategy of identifying a good ligand that fits the protein's binding site both geometrically and energetically, is an effective and competent method that is becoming increasingly important in logical drug design [20]. The drug research business uses a variety of molecular docking software. The most widely used and well-liked molecular docking software programs are AutoDock [13–15], AutoDock and Schrodinger. In accordance with the work done to find new AChE inhibitors, we conduct a comparative docking analysis using two widely used programs: AutoDock and Schrodinger Glide tool. By docking 65 ligands with AChE protein and comparing the expected binding affinities with the observed values, the docking accuracy of the chosen docking techniques were assessed.
Numerous plant species contain coumarin, a naturally occurring heterocyclic molecule that can be created by synthetic processes. Many heteroaromatic compounds and derivatives, especially those containing nitrogen atoms, were created in order to investigate their AChE/BuChE inhibitory potential for treating AD. Fig. 1 provides a clear explanation of the designing technique. This study set out to determine the lead molecules (synthesized 7-hydroxycoumarin derivatives) and investigate the viability of two distinct docking strategies for our target AChE: AutoDock vina and Schrodinger Glide. Using these tools, we have conducted a comparative analysis of ligand binding affinity and binding poses.
MATERIALS AND METHODS
Molecular docking studies of designed derivatives
A lot of ligands' binding affinities can be predicted using molecular docking techniques. The purpose we had in this work was to investigate whether there might already be a connection between the docking scores and the experimental bioactivities of the inhibitors being studied, as well as to compare the 2D interaction between the protein and the ligand exhibited by Schrodinger and AutoDock vina. Using the above designing strategy, 65 coumarin derivatives were designed, out of which the best 15(4a-o) were synthesized and evaluated for their anti-Alzheimer activity, which was explained in our previous work [21]. The detailed docking studies had been performed to get a complete insight into the molecular interactions of designed compounds with three-dimensional hAChE (PDB code: 4EY7). Using the Glide module of Schrodinger Maestro 2018-1 and Auto Dock vina, molecular docking research was conducted to evaluate the binding affinity and pose stability of compounds (4a-4o) in the active sites of hAChE (PDB Code: 4EY7). The protein's crystalline structure was generated.
Initially, partial charges were provided after hydrogens were supplied. After optimizing the protein structure at pH 7.0 using the PROPKA technique, the RMSD of the convergence heavy atoms was maintained at 0.30Å during constrained minimization. The receptor grid was formed in order to identify active sites that were less than 10× 10× 10 Å from the centroid of the co-crystallized ligand (donepezil) [22]. Molecular docking analysis is used to elucidate donepezil’s manner of binding with the receptor proteins using Glide.
Similarly, AutoDock Tools (ADT) was used to finish intermediate processes, including creating grid boxes and preparing pdbqt files for proteins and ligands. Polar hydrogens, unified atom Kollman charges, and solvation parameters were assigned using ADT and protein fragmental volumes. The prepared file was saved by AutoDock in PDBQT format. Using a grid box, Auto Grid was utilized to create the grid map. The grid center was identified with dimensions (x, y, and z) of -1.095,-1.554, and 3.894. The grid size was set to 60 × 60 × 60 xyz points with grid spacing of 0.375 Å. The ligand structure is used to compute a score grid, which reduces computation time. For additional study, the position with the lowest binding energy or affinity was taken out and matched with the receptor structure [23, 24]. Using the AutoDock tool, molecular docking analysis is utilized to clarify how donepezil binds to the receptor proteins.
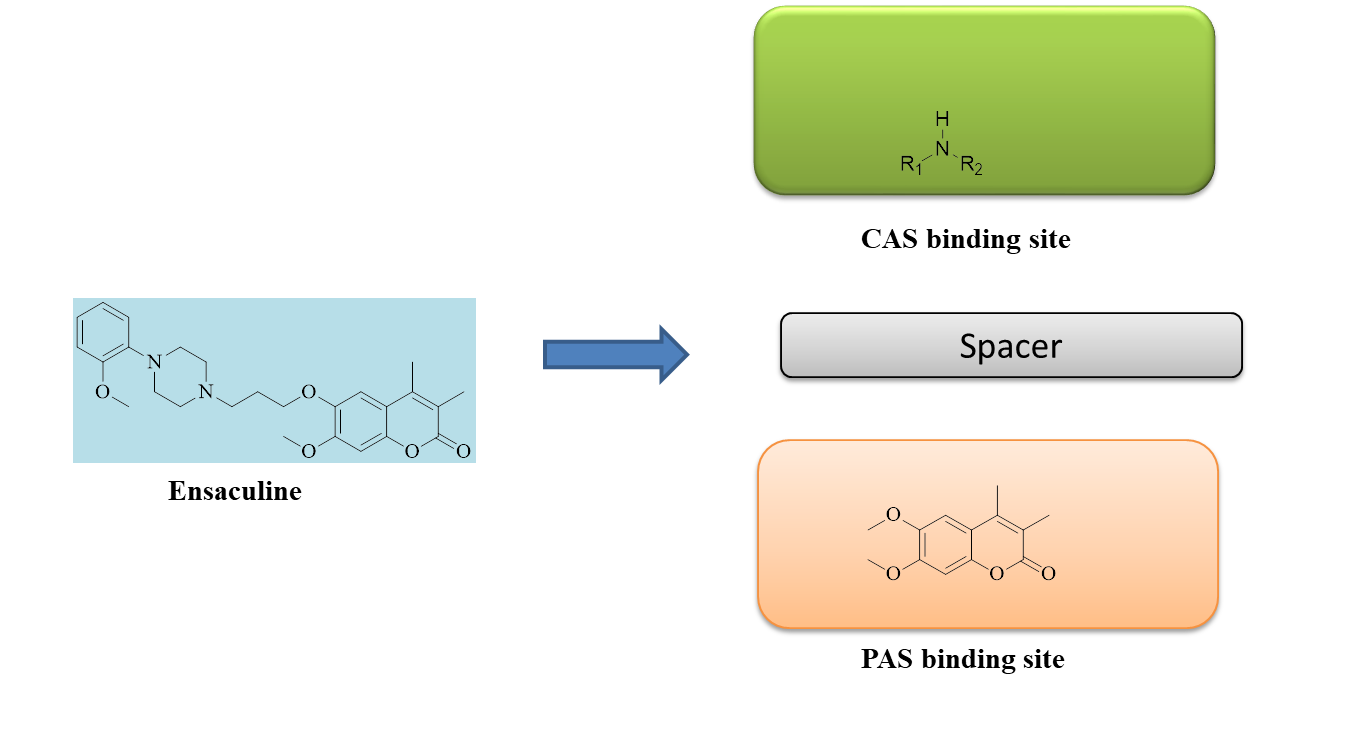
Fig. 1: Design strategy for 7-hydroxy coumarin analogues
RESULTS AND DISCUSSION
This work compares two popular docking techniques (AutoDock and Schrodinger Glide) in order to determine an accurate docking routine for performing molecular docking studies of the AChE protein and to find the strongest inhibitors against that protein. The binding affinities obtained with both the tools are given in table 1. The best ten docked poses for each ligand were returned by each docking procedure. Table 2 provides an overview of the quantity and classifications of the dock poses. Schrodinger was determined to be the most effective of the two docking methods that were examined for both docking and creating postures that bind most effectively deep within the binding pocket's 5 Å. 84% of the poses it produced were good. Autodock, however, did well as well, striking 68% of good postures. Docking programs must operate at about equal speeds in order to be evaluated for docking accuracy. Additionally, compared is the average amount of time needed to complete the docking calculation for a single ligand using various scoring systems and docking procedures. Glide is the fastest algorithm in terms of the average time needed to dock a single ligand. Fig. 2 and 3 shows the binding interactions of compounds in bar graph form.
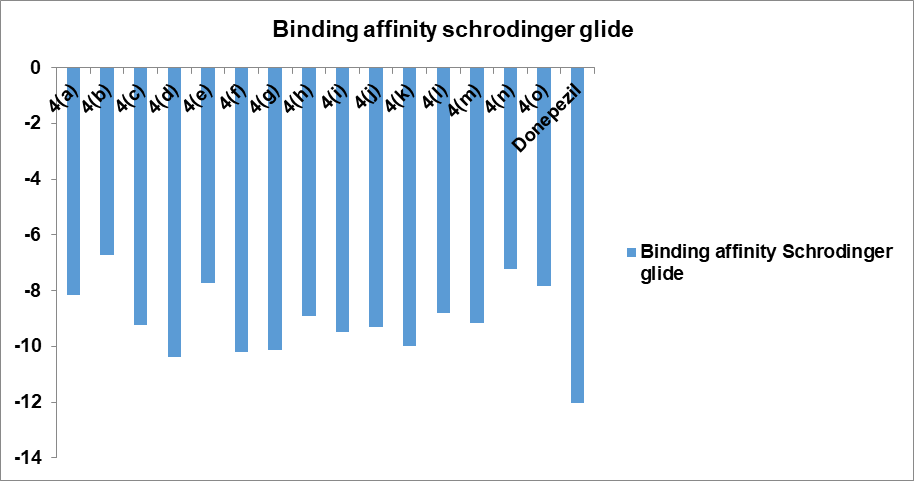
Fig. 2: Graph of docking score of 7-hydroxy coumarin derivatives
Table 1: Docking score of 7-hydroxy coumarin derivatives
| S. No. | Compound code | Compound structure | Binding affinity Schrodinger Glide | Binding affinity Autodock Vina |
| 1. | 4(a) |  |
-8.148 | -6.267 |
| 2. | 4(b) |  |
-6.708 | -5.907 |
| 3. | 4(c) | 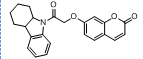 |
-9.222 | -7.435 |
| 4. | 4(d) | 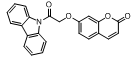 |
-10.384 | -8.342 |
| 5. | 4(e) | 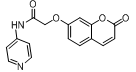 |
-7.722 | -6.733 |
| 6. | 4(f) |  |
-10.211 | -8.222 |
| 7. | 4(g) |  |
-10.142 | -6.765 |
| 8. | 4(h) |  |
-8.902 | -6.892 |
| 9. | 4(i) | 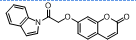 |
-9.482 | -7.115 |
| 10. | 4(j) | 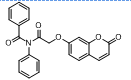 |
-9.322 | -6.563 |
| 11. | 4(k) | 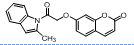 |
-9.992 | -7.093 |
| 12. | 4(l) | 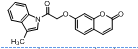 |
-8.802 | -6.942 |
| 13. | 4(m) | 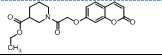 |
-9.162 | -6.531 |
| 14. | 4(n) |  |
-7.212 | -5.985 |
| 15. | 4(o) | 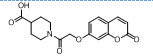 |
-7.824 | -5.467 |
| 16. | Donepezil |  |
-12.04 | -12.04 |
Table 2: Number of good, fair and poor docked complexes obtained by both docking routines
| Pose | Binding affinity Schrodinger Glide | Binding affinity Autodock Vina |
| Good | 55 | 44 |
| Fair | 06 | 13 |
| Poor | 04 | 08 |
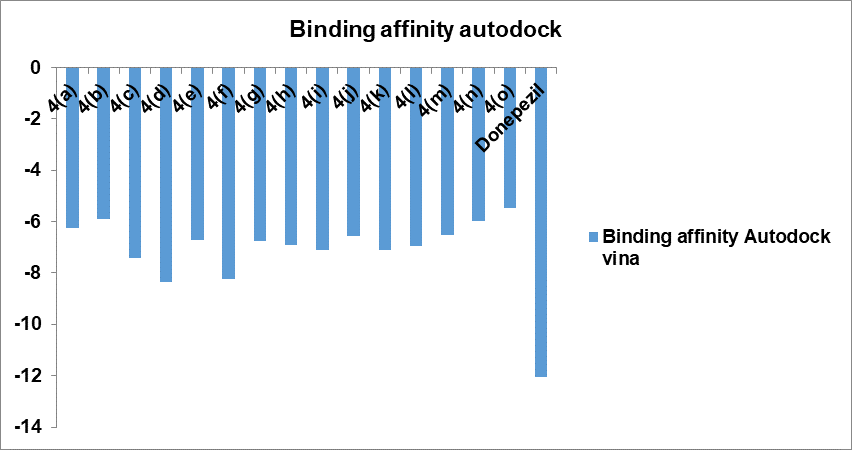
Fig. 3: Graph of docking score of 7-hydroxy coumarin derivatives

Fig. 4a: Binding interactions of donepezil with enzyme with glide
Molecular docking studies of donepezil and important coumarin analogues
Below fig. displays the 2D depiction of protein–ligand interactions in the optimal postures produced by both programs under study. Every molecule has the same binding mode, as shown in the figure. Remarkably, these atoms and the residues have significant interactions that directly contribute to this enzyme's catalytic function. The binding interactions of donepezil with the enzyme are shown with AutoDock vina (fig. 4a) and Glide (fig. 4b). The primary stabilizing factors for the ligand–enzyme complexes are hydrophobic and hydrogen bonding interactions. Each docking technique produced a top docked pose that showed formed bonds with one or more amino acids in the AChE binding pocket. In the majority of docking applications, the top-ranked pose with the lowest docked binding affinities and the highest docking scores is typically chosen as the default.
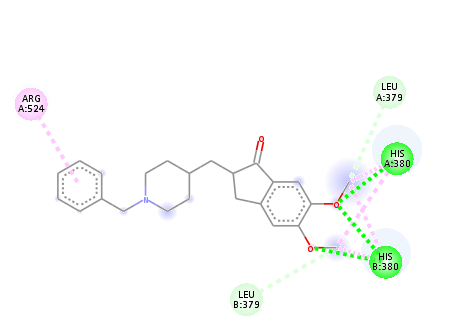
Fig. 4b: Binding interactions of donepezil with enzyme with AutoDock vina
Molecular docking studies of the first most active compound 4 (d)
As before, a molecular docking analysis was performed using one of the most active compounds, 4 (d), to clarify the inhibitor's mode of binding. Compound 4 (d) and donepezil had docked scores of-10.384 and-12.040, respectively. The interaction between compound 4(d) in its ideal docking position and the amino acids in the hAchE active site is shown in fig. 5. In the catalytic triad of the CAS region, the carbazole ring established a polar contact with HIS 447 and SER 203. The carbazole ring displayed hydrophobic contacts with PHE 338 and TYR 337, pi-pi stacking with TRP 86, and electrostatic interactions with GLU 202 at the anionic subsite. At the acyl binding pocket, the coumarin moiety's carbonyl group established a hydrogen connection with PHE 295 and a hydrophobic contact with PHE 297. Compound 4(d) reacted with glycine residues (GLY 121 and GLY 122) at the oxyanion site. In the PAS region, the coumarin moiety demonstrated hydrophobic contacts with TYR 72 and TYR 341 and pi-pi stacking with amino acid TRP 286. It also shows electrostatic interactions with ASP 74. Furthermore, in PAS, the carbonyl group of the amide linkage demonstrated hydrogen bonding with TYR 124.

Fig. 5: 2D Interactions between hAChE and compound 4(d) by glide
Fig. 6 depicts the interaction between protein and the compound 4(d) shown by AutoDock tool. The all the three rings of carbazole formed ionic interaction or salt bridge formation with HIS 381 in the catalytic triad of the CAS region. Both the rings of coumarin moiety forms pi-pi interaction with ALA 397. Moreover, Ligand shows vander waals interactions with GLU 396, TYR 382, THR 75, ARG 525, LEU 380 amino acids in binding pockets.
Molecular docking studies of the second most active compound, 4(f)
Compound 4 (f) and donepezil had docked scores of-10.211 and-12.040 kcal/mole, respectively. The interaction between compound 4 (f) in its ideal docking position and the amino acids in the hAchE active site is shown in fig. 7. In the catalytic triad of the CAS area, the pyrone ring of coumarin produced pi-pi stacking and polar contacts with HID 447, while the carbonyl group of the pyrone ring made a hydrogen bond with SER 203. The coumarin's benzene ring displayed pi-pi stacking with TYR 337 at the anionic subsite. Furthermore, in the anionic subsite, compound 4 (f) demonstrated hydrophobic contacts with PHE 338 and electrostatic interactions with GLU 202. Compound 4(f)'s amide linkage's carbonyl group established a hydrogen bond with PHE 295 and a hydrophobic interaction with PHE 297 at the acyl binding pocket. Compound 4(f) developed a hydrophobic contact with ALA 204 at the oxyanion site after interacting with glycine residues (GLY 121 and GLY 122).
Additionally, compound 4 (f) demonstrated hydrophobic contacts with TRP 286, TYR 124, and TYR 72, as well as electrostatic interactions with ASP 74 in the PAS region. The coumarin moiety's benzene ring also displayed pi-pi stacking with amino acid TYR 341. One interesting characteristic of AChE inhibitors is its ability to bind with both enzyme sites, as demonstrated in compound 4(f).
Fig. 8 depicts the interaction between hAchE and the compound 4(d) shown by AutoDock tool. The carbonyl group at the coumarin moiety forms a conventional hydrogen bond with ALA 526. Both the rings of coumarin forms pi-pi interactions with ALA 528 and ARG 525. Moreover, the ligand shows Vander Waals interactions with HIS 381, LEU 386, ARG 522, GLN 527 and TYR 382 amino acids in binding pockets.
Docking interactions of other important derivatives
2D images of the active site interactions between hAChE and some of the other compounds as studied with both tools, are summarized below.

Fig. 6: 2D interactions between hAChE and compound 4(d) by AutoDock
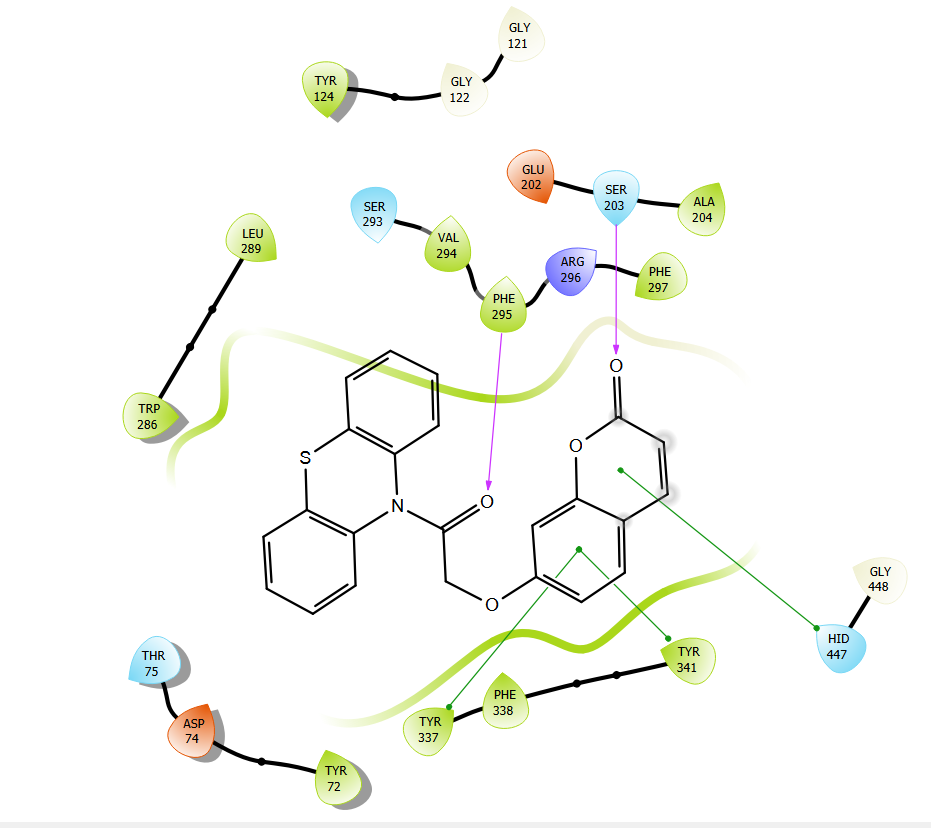
Fig. 7: 2D Interactions between hache and compound 4(f) by glide

Fig. 8: 2D Interactions between hAChE and compound 4(f) by AutoDock
Compound 4 (a)
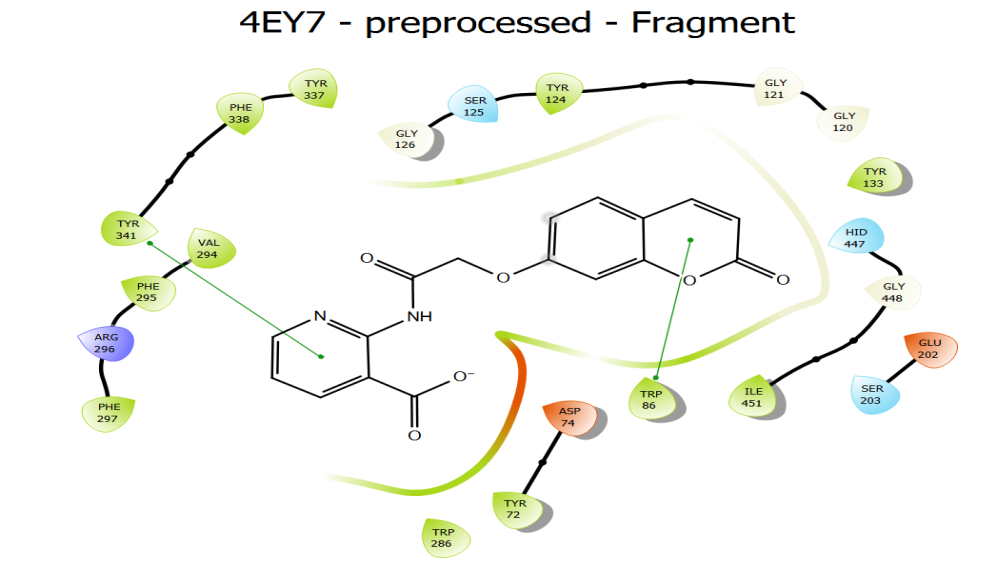
Fig. 9a: 2D interactions of compound 4 (a) with glide
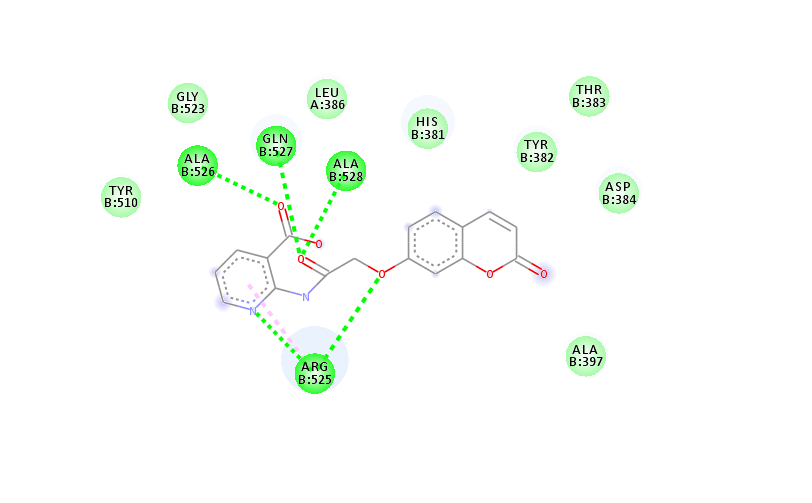
Fig. 9b: 2D interactions of compound 4 (a) with AutoDock
Compound4 (b)
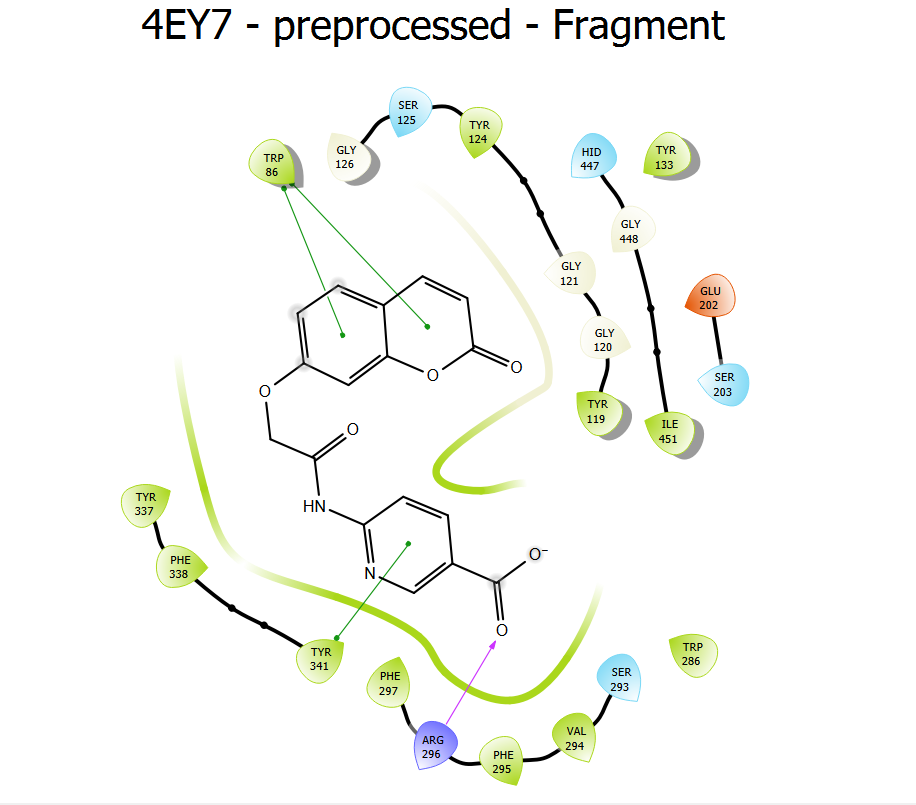
Fig. 9c: 2D interactions of compound 4 (b) with glide
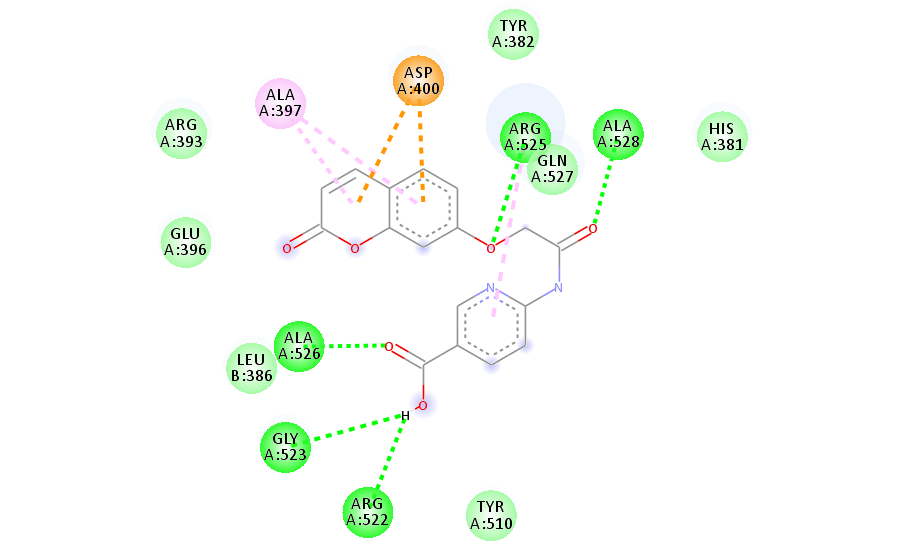
Fig. 9d: 2D interactions of compound 4 (b) with AutoDock
Compound4(c)

Fig. 9e: 2D interactions of compound 4 (c) with glide

Fig. 9f: 2D interactions of compound 4 (c) with AutoDock
Compound4 (h)

Fig. 9g: 2D interactions of compound 4 (h) with glide
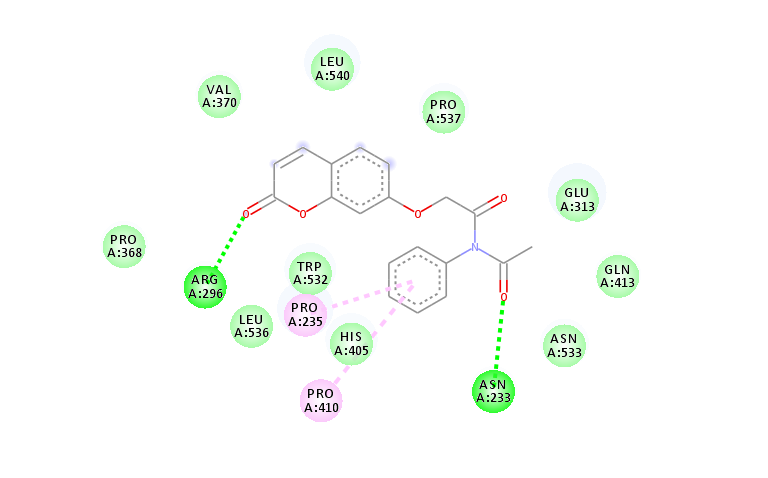
Fig. 9h: 2D interactions of compound 4 (h) with AutoDock
Compound4 (m)
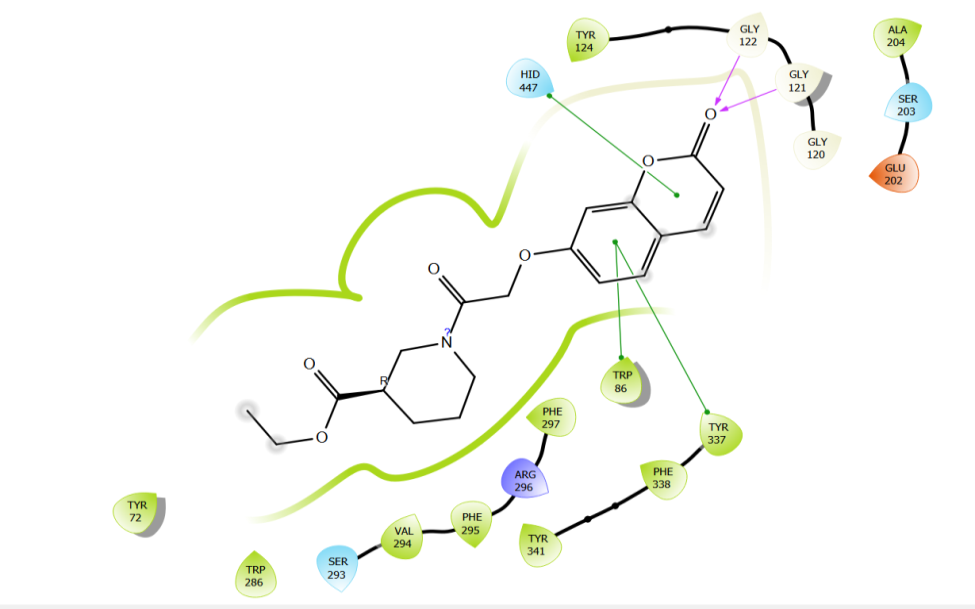
Fig. 9i: 2D interactions of compound 4 (m) with glide
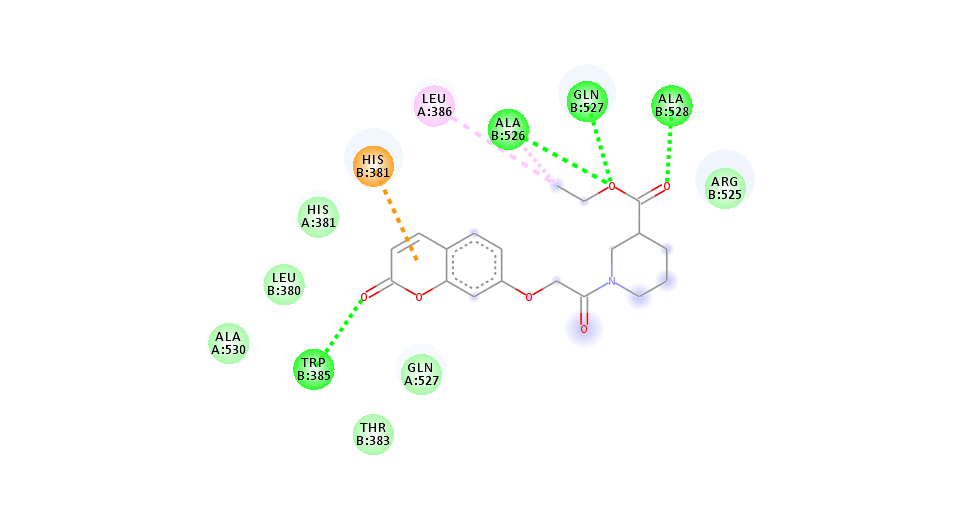
Fig. 9j: 2D interactions of compound 4 (m) with AutoDock
Based on their docking score, all drugs demonstrated similar binding to AChE (in comparison to donepezil). Docking scores for compounds were in the range-5.467 to-8.342 using Auto dock and -10.384 to-6.708 using Glide. Donepezil binds to both CAS and PAS sites of the enzyme. The compounds 4(d), 7-(2-(9H-carbazol-9-yl)-2-oxoethoxy)-2H-chromen-2-one and 4(f), 7-(2-oxo-2-(10H-phenothiazin-10-yl) ethoxy)-2H-chromen-2-one having the highest docking scores using both the tools. All the compounds also displayed more or less the same pi-pi stacking interactions, hydrophobic interactions, polar interactions etc. Molecular docking results revealed that the compounds 4 (d) and 4 (f) possess acetylcholinesterase inhibitory properties by binding to both CAS and PAS in the active sites of AChE. It is also supported by previously published articles that the coumarin derivatives exhibit anticholinesterase activity by dual binding mode in the active sites of AChE [25]. Moreover, the compounds showing dual binding site interactions as Donepezil, at molecular level of inhibitory activity, are akin to exhibit better acetylcholinesterase inhibitory properties [26].
5. A comprehensive review of the molecular docking interactions of compounds 4 (d), 4 (f) and donepezil in hAChE (4EY7)
Table 3: Molecular docking Interaction of compound 4 (d), 4 (f) and donepezil in hAChE (4EY7)
| Compd | Schrodinger Interacting residues | AutoDock vina | |||
| H-bonds | Hydrophilic bonds | Remaining residues | Pi-alkyl/pi-pi stalked/ Hydrophobic interactions | Remaining residues | |
| 4 (d) | HIS 447 SER 203 | THR 75, GLY120 | TRP 286, TYR 72, TYR 341, ASP 74, TYR 124 | ALA 397,HIS 381 | GLU 396,ARG 393,TYR 382,HIS 381, THR 383,ASP 400,ASP 384 |
| 4 (f) | HID 447 SER 203 | SER 293, THR 75,ARG 296, GLY 448 |
TYR 341, TRP 286, TYR 124, TYR 72, ASP 74 | ALA 526 ALA 528 ARG 525 |
GLY 523,ARG 522,ARG 525,HIS 381,TYR 382 |
| Donepezil | HIS 447 SER 239,SER 203 | PHE 295, PHE 297 | TRP 286, TYR 341, TYR 124, TYR 72, ASP 74. | ARG 524,HIS 380 | LEU 379 |
CONCLUSION
Over the past few years, docking and scoring have undergone major changes. It is now a useful tool in the process of finding new drugs. Using AutoDock and Schrodinger tools, we have conducted a comparative analysis of ligand binding affinity. Schrodinger was found to be comparatively more helpful in blind docking pose prediction and consistently outperformed another program. The ligand docking experiments demonstrated that the amino acid residues are involved in the binding pocket. Donepezil was used to compare the docking score and interactions of designed compounds with amino acid residues in of hAChE. hydrogen bond formation and electrostatic interaction akin to those of donepezil. Molecular docking results revealed that the compounds possess acetylcholinesterase inhibitory properties by binding to both CAS and PAS in the active sites of AChE.
ACKNOWLEDGMENT
The authors are thankful to Banasthali Vidyapith, Jaipur, Rajasthan, India, for their support.
FUNDING
No organization provided funding to the authors for the submitted work.
AUTHORS CONTRIBUTIONS
Dr. Tyagi Alka has done drafting, designing, writing and all the work related to the manuscript. Ms. Sadgir Priyanka has reviewed the work.
CONFLICT OF INTERESTS
No conflicts of interest are disclosed by the authors
REFERENCES
Dorababu A. Promising heterocycle-based scaffolds in recent (2019-2021) anti-Alzheimer’s drug design and discovery. Eur J Pharmacol. 2022 Apr 5;920:174847. doi: 10.1016/j.ejphar.2022.174847, PMID 35218718.
Kulisevsky J. Role of dopamine in learning and memory: implications for the treatment of cognitive dysfunction in patients with Parkinson’s disease. Drugs Aging. 2000;16(5):365-79. doi: 10.2165/00002512-200016050-00006, PMID 10917074.
Sharma P, Srivastava P, Seth A, Tripathi PN, Banerjee AG, Shrivastava SK. Comprehensive review of mechanisms of pathogenesis involved in Alzheimer’s disease and potential therapeutic strategies. Prog Neurobiol. 2019 Mar;174:53-89. doi: 10.1016/j.pneurobio.2018.12.006, PMID 30599179.
Tyler CR, Allan AM. The effects of arsenic exposure on neurological and cognitive dysfunction in human and rodent studies: a review. Curr Environ Health Rep. 2014;1(2):132-47. doi: 10.1007/s40572-014-0012-1, PMID 24860722.
Choubdar N, Golshani M, Jalili Baleh L, Nadri H, Kucukkilinc TT, Ayazgok B. New classes of carbazoles as potential multi-functional anti-Alzheimer’s agents. Bioorg Chem. 2019 Oct;91:103164. doi: 10.1016/j.bioorg.2019.103164, PMID 31398601.
Rani P, Singh K, Arjuna A, Devi S. Genetic disorder Alzheimer. Asian J Pharm Clin Res. 2017;10(12):36-9. doi: 10.22159/ajpcr.2017.v10i12.18684.
Tham W, Auchus AP, Thong M, Goh ML, Chang HM, Wong MC. Progression of cognitive impairment after stroke: one-year results from a longitudinal study of singaporean stroke patients. J Neurol Sci. 2002;203-204(6):49-52. doi: 10.1016/s0022-510x(02)00260-5, PMID 12417356.
Tripathi PN, Srivastava P, Sharma P, Tripathi MK, Seth A, Tripathi A. Biphenyl-3-oxo-1,2,4-triazine-linked piperazine derivatives as potential cholinesterase inhibitors with anti-oxidant property to improve the learning and memory. Bioorg Chem. 2019 Apr;85:82-96. doi: 10.1016/j.bioorg.2018.12.017, PMID 30605887.
Piplani P, Danta CC. Design and synthesis of newer potential 4-(N-acetylamino) phenol-derived piperazine derivatives as potential cognition enhancers. Bioorg Chem. 2015 Jun;60:64-73. doi: 10.1016/j.bioorg.2015.04.004, PMID 25965977.
Sharma V. Alzheimer’s disease: a consequence of impaired mitophagy? Asian J Pharm Clin Res. 2019;12(2):75-80. doi: 10.22159/ajpcr.2019.v12i2.28407.
Korolev IO. Alzheimer’s disease: a clinical and basic science review. Med Student Res J. 2014 Sep;4:24-33.
Yusufzai SK, Khan MS, Sulaiman O, Osman H, Lamjin DN. Molecular docking studies of coumarin hybrids as potential acetylcholinesterase, butyrylcholinesterase, monoamine oxidase A/B and β-amyloid inhibitors for Alzheimer’s disease. Chem Cent J. 2018;12(1):128. doi: 10.1186/s13065-018-0497-z, PMID 30515636.
Sirvio J, Riekkinen P, Jakala P, Riekkinen PJ. Experimental studies on the role of serotonin in cognition. Prog Neurobiol. 1994;43(4-5):363-79. doi: 10.1016/0301-0082(94)90060-4, PMID 7816931.
Blokland A. Acetylcholine: a neurotransmitter for learning and memory? Brain Res Brain Res Rev. 1995;21(3):285-300. doi: 10.1016/0165-0173(95)00016-x, PMID 8806017.
Shi DH, Min W, Song M, Si XX, Li MC, Zhang Z. Synthesis, characterization crystal structure and evaluation of four carbazole coumarin hybrids as multifunctional agents for the treatment of Alzheimer’s disease. J Mol Struct. 2020 Jun 5;1209:127897. doi: 10.1016/j.molstruc.2020.127897.
Hu YH, Yang J, Zhang Y, Liu KC, Liu T, Sun J. Synthesis and biological evaluation of 3-(4-aminophenyl)-coumarin derivatives as potential anti-Alzheimer’s disease agents. J Enzyme Inhib Med Chem. 2019;34(1):1083-92. doi: 10.1080/14756366.2019.1615484, PMID 31117844.
Piazzi L, Cavalli A, Colizzi F, Belluti F, Bartolini M, Mancini F. Multi-target-directed coumarin derivatives: hAChE and BACE1 inhibitors as potential anti-Alzheimer compounds. Bioorg Med Chem Lett. 2008;18(1):423-6. doi: 10.1016/j.bmcl.2007.09.100, PMID 17998161.
Makhaeva GF, Shevtsova EF, Boltneva NP, Lushchekina SV, Kovaleva NV, Rudakova EV. Overview of novel multifunctional agents based on conjugates of γ-carbolines carbazoles tetrahydrocarbazoles, phenothiazines and aminoadamantanes for treatment of Alzheimer’s disease. Chem Biol Interact. 2019 May;308:224-34. doi: 10.1016/j.cbi.2019.05.020, PMID 31100279.
Zheng H, Fridkin M, Youdim M. From single target to multitarget/network therapeutics in Alzheimer’s therapy. Pharmaceuticals (Basel). 2014;7(2):113-35. doi: 10.3390/ph7020113, PMID 24463342.
Terry RD, Gonatas NK, Weiss M. Ultrastructural studies in Alzheimer’s presenile dementia. Am J Pathol. 1964;44(2):269-97. PMID 14119171.
Walhekar VM, Bathe V, Kadam V, Rokade S. Review on Embelia ribes and it’s pharmacological activity. International Journal of Pharmaceutical Sciences. 2024;2(4):1312-9. doi: 10.5281/zenodo.11092906.
Bhawana S, Alka T, Anurag. Design creation and biological screening of newer coumarin-coupled heterocyclic hybrids as acetylcholinesterase inhibitors that may be useful in the treatment of Alzheimer’s disease. Pharm Chem J. 2024;58(7):1069-83. doi: 10.1007/s11094-024-03245-4.
Kono Y. Generation of superoxide radical during autoxidation of hydroxylamine and an assay for superoxide dismutase. Arch Biochem Biophys. Science Direct. 1978;186(1):189-95. doi: 10.1016/0003-9861(78)90479-4, PMID 24422.
Azam SS, Abbasi SW. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor Biol Med Model. 2013 Oct 24;10:63. doi: 10.1186/1742-4682-10-63, PMID 24156411.
Sharma P, Tripathi A, Tripathi PN, Singh SS, Singh SP, Shrivastava SK. Novel molecular hybrids of N-Benzylpiperidine and 1,3,4-oxadiazole as multitargeted therapeutics to treat Alzheimer’s disease. ACS Chem Neurosci. 2019;10(10):4361-84. doi: 10.1021/acschemneuro.9b00430, PMID 31491074.
Silva MA, Kiametis AS, Treptow W. Donepezil inhibits acetylcholinesterase via multiple binding modes at room temperature. J Chem Inf Model. 2020;60(7):3463-71. doi: 10.1021/acs.jcim.9b01073, PMID 32096991.